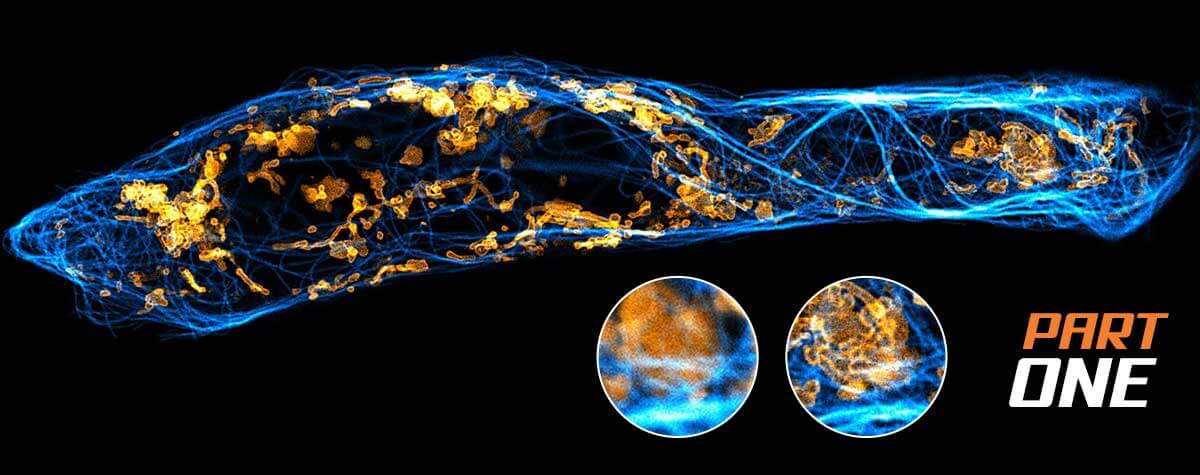
Superresolution for biology:
The spatial resolution achievable with today’s light microscopes has unveiled life at the scale of individual molecules. Size is no longer a barrier to seeing biology at the most fundamental level. But life is not static. It emerges from movement and change. How do superresolution technologies hold up to the challenges of documenting dynamic biological mechanisms?
when size, time, and context matter
Superresolution matters in biology
Biological research has penetrated deep into the realm of the smallest. Today, we study individual cells rather than average their behavior across tissues. We identify unknown microbial species based on novel DNA sequences alone. We’ve even explored quantum underpinnings of photosynthesis and the magnetic compass of migrating birds. The resolution with which we see life has made leaps and bounds, in no small part due to advances in microscopy. But when does superresolution have the greatest impact on how we understand life?
Biology as a science of the smallest
Fascinating biology takes place at scales invisible to the naked eye. And with molecular biology driving this century’s boon in biotechnology and biomedicine, it is no understatement to say that superresolution matters in biology. Fact is that the interactions and processes of life transpire at scales below the diffraction barrier.
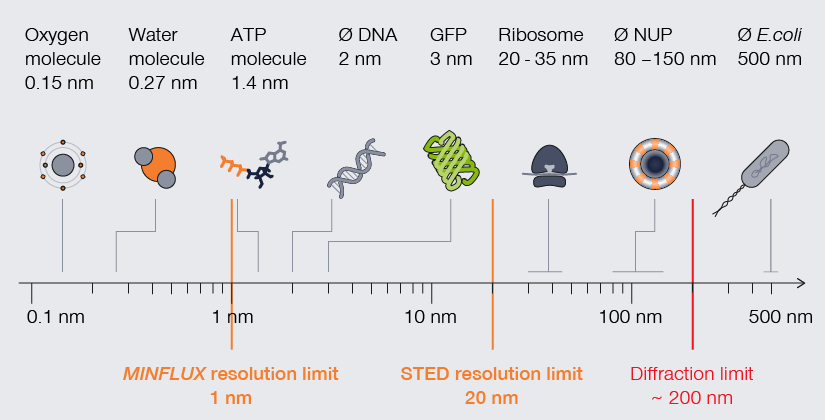
The scale of life. High-resolution understanding of life happens below the diffraction barrier.
A range of light microscopy technologies reach and surpass the light diffraction barrier of about 200 nm, among them stimulated emission depletion (STED) and MINFLUX. As methods, they vary widely in complexity of setup, sample preparation, data processing after image capture, and capacity for live-specimen imaging. Together, however, they grant access to the nanoscale world increasingly nearing the resolution of an electron microscope. Thus, today a biologist who needs to visualize a cellular structure or molecule in the low nanometer scale can contemplate numerous alternatives to highly specialized methods that require expertise, specialized equipment, time, and sample destruction. As we’ll see, those alternatives are essential to the exploration of biology in more ways than one.
When size matters
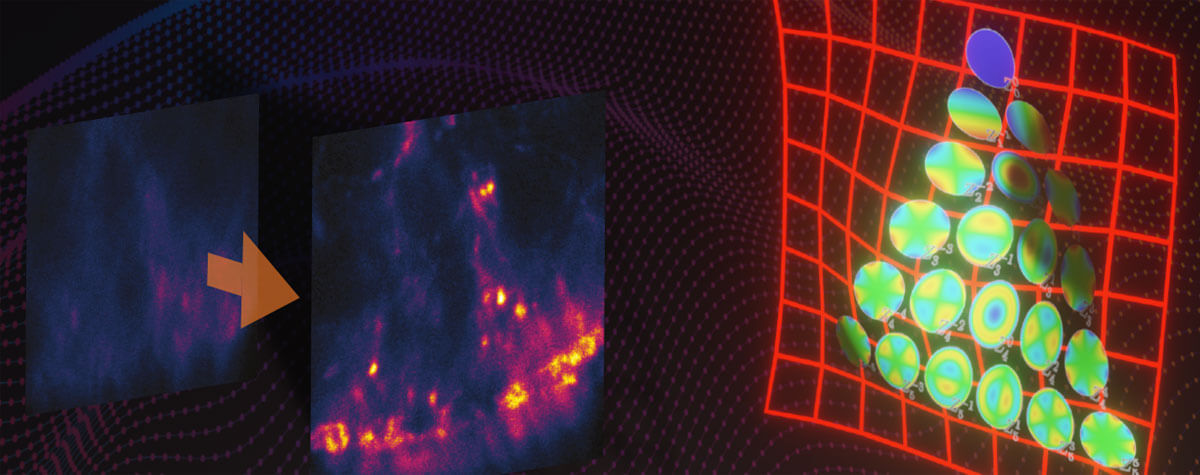
For decades, electron microscopy dominated with mind-blowing images of subcellular components and molecules. Now however, superresolution technologies make that detailed imagery standard in light microscopy. Consider mitochondria. The folds of their inner membrane – or cristae – is where they synthesize ATP. Mitochondria and their cristae are highly dynamic, adjusting in shape and quantity. Visualizing cristae can help characterize how those changes correlate with the metabolic state of a cell but requires a resolution far below the average distance between cristae (~100 nm). Using newly developed fluorescent dyes, researchers have applied stimulated emission depletion (STED) microscopy to image the characteristic stripes of mitochondria at resolutions as high as 35 nm. With MINFLUX, resolutions as high as 2 nm can be reached, which allows to tell apart individual molecules.
When time matters
Size and distance measurements are important to characterize subcellular environments, but with them alone we cannot unravel life in all its facets. Life is about movement and interactions, and the timescales at which those happen span from nanoseconds for protein conformational changes to minutes for DNA replication. Clearly, the temporal resolution of imaging is decisive in making meaningful biological observations at the molecular level. Electron microscopy meets its limits here as samples need to be fixed and cannot provide any information about temporal correlations.
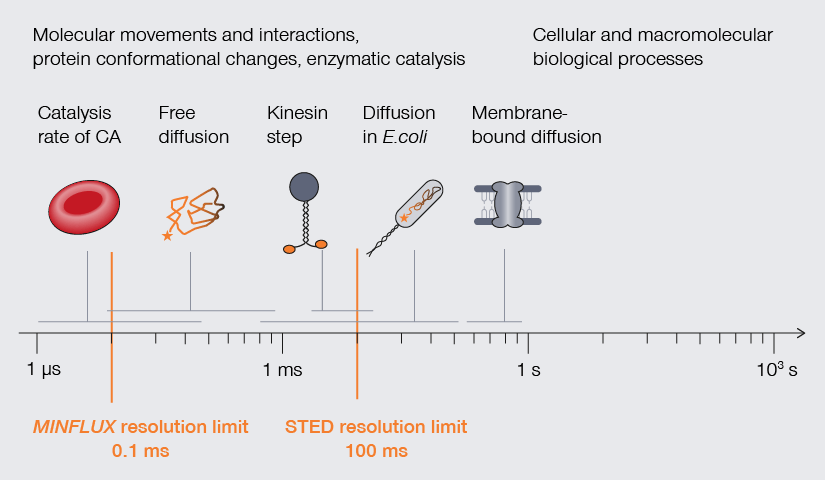
Temporal resolution is crucial to understand molecular movements and interactions, protein conformational changes, and enzymatic catalysis.
A handful of superresolution methods accommodate the temporal resolution required to observe processes of life (Fig. 2). STED, for example, offers fast imaging speed (roughly 1 frame per second). Using photostable dyes that endure long-term imaging, Yang and colleagues documented changes in the shape of individual mitochondria over a 10-minute and a 60-minute timeframe. The movies show the formation of bubbles in mitochondria undergoing fission. Those bubbles fill rapidly with cristae and then separate to form individual small mitochondria.1 Other studies have used superresolution methods to track proteins, like the movement of AMPA receptors in neurons, the translocation of membrane-bound glycophosphatidylinositol (GPI) molecules, and even the diffusion of Hfq protein as it binds mRNA in E. coli. Finally, MINFLUX takes it to the next level: Thanks to its photon efficiency, it can resolve molecular movements with a temporal resolution in the range of 100 µs while providing highest spatial detail, making it possible to track individual lipids diffusing in biological membranes or the conformational changes of kinesin-1 as it walks along microtubules. 3,4,5
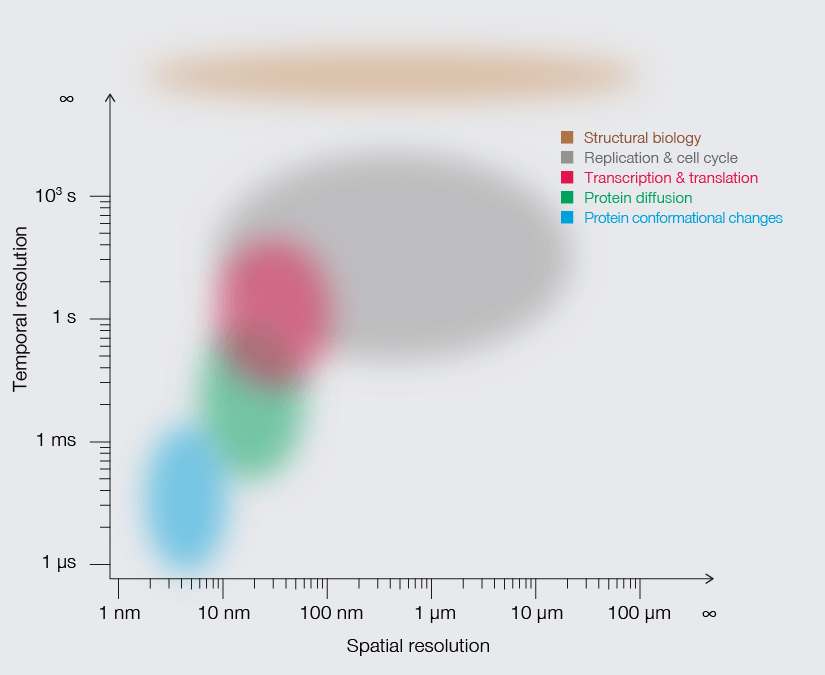
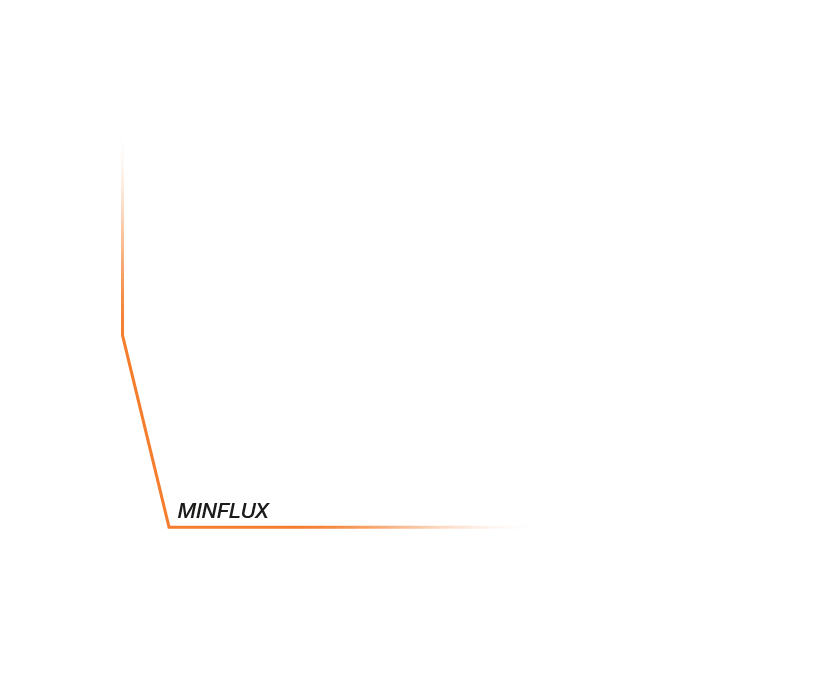
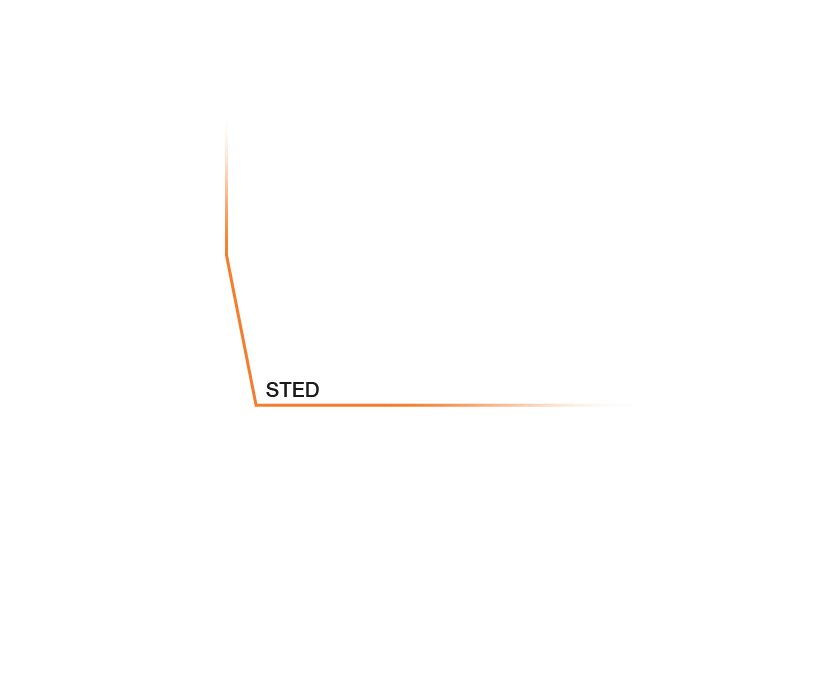
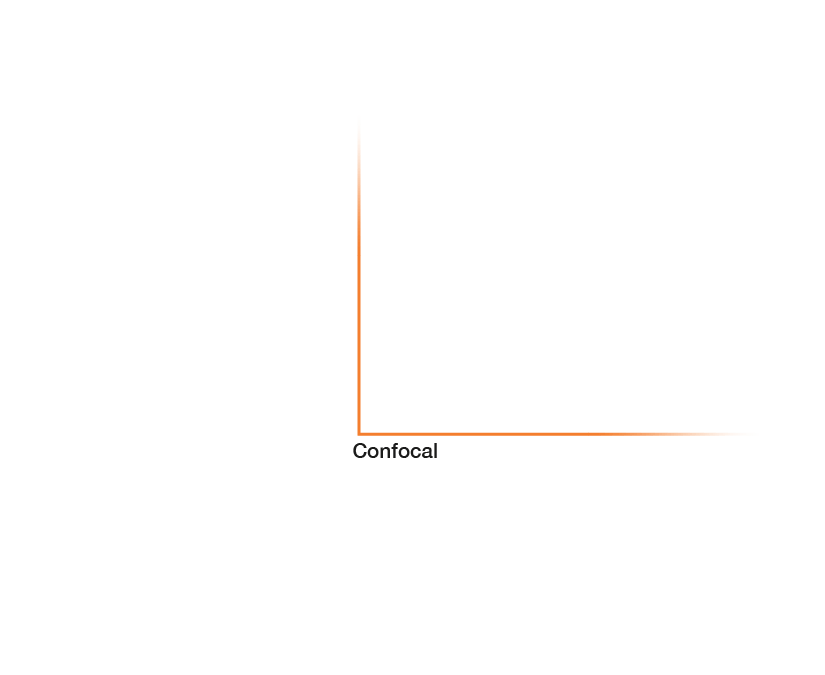
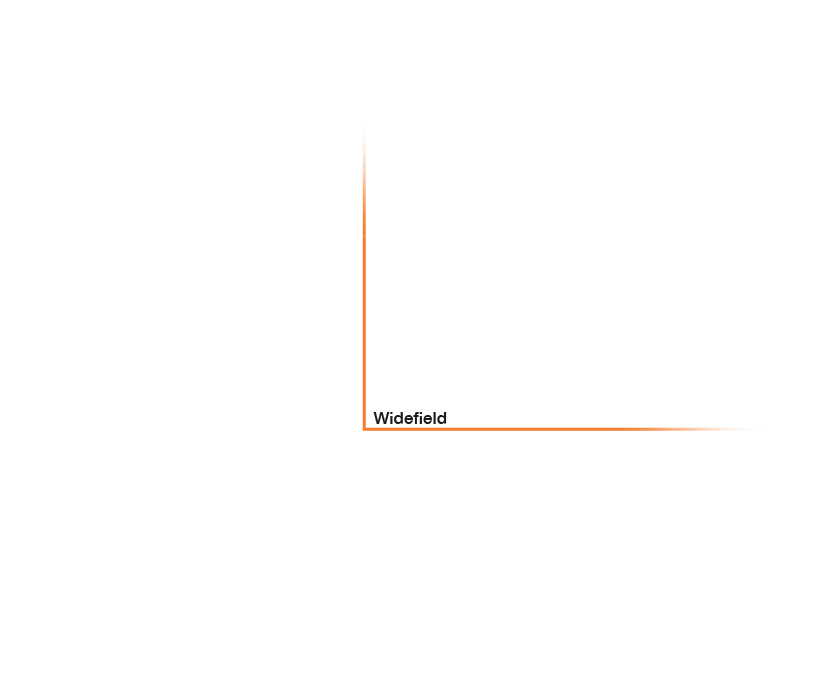


Approximate temporal and spatial resolution range of microscopy methods.
When context matters
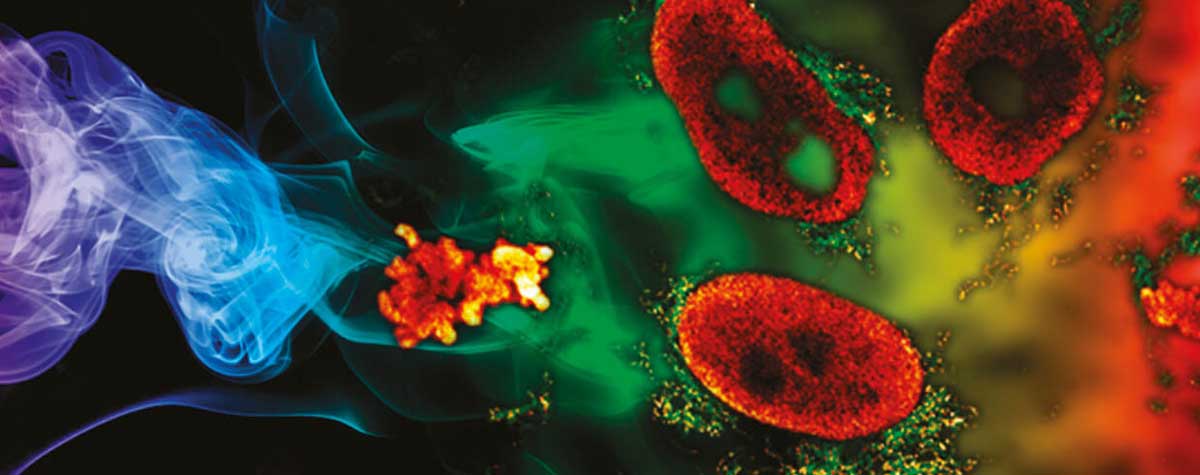
The long-term imaging of biological processes at high resolution is not just about imaging speed. It’s also about keeping your sample alive. That is, after all, the context of biological research. The difficulty here lies in preventing specimen destruction through preparation techniques or the toxic effects of exposing live cells to high-energy or lengthy excitation cycles. Creative solutions to this problem can be found in the literature. For example, event-triggered STED limits photobleaching and toxicity by linking automated STED imaging to the detection of a cellular event, like protein recruitment or vesicle trafficking. 6 In a particular exciting demonstration of live imaging, STED microscopy was used to record dynamic changes in dendritic spines of cortical somatosensory neurons in a mouse. The mouse was alive during the procedure and the fine details of these neural structures were revealed with a spatial resolution of under 70 nm.7
Superresolution in biology is so much more than size
Looking into the future, our understanding of biological processes to leverage them for applications will increasingly rely on methods that offer more than exceptional spatial resolution. They must also match the time scale of biological events and capture molecular interactions in vivo. Advances in superresolution microscopy are at the cusp of unifying those three dimensions into one easy-to-use technology. Our view of life is looking clearer than ever before.
1 Yang, X. et al. 2022. Mitochondrial dynamics quantitatively revealed by STED nanoscopy with an enhanced squaraine variant probe. Nature Communications, 11:3699. DOI: 0.1038/s41467-020-17546-1
2 Turkowyd, B. et al. 2016. From single molecules to life: microscopy at the nanoscale. Anal Bioanal Chem. 408:6885. DOI: 10.1007/s00216-016-9781-8
3 Schmidt et al. 2021. MINFLUX nanometer-scale 3D imaging and microsecond-range tracking on a common fluorescence microscope. Nature Communications 12: 1478. https://www.nature.com/articles/s41467-021-21652-z
4 Wolff, J. O. et al. MINFLUX dissects the unimpeded walking of kinesin-1. Science 379, 1004–1010 (2023). DOI: 10.1126/science.ade2650
5 Deguchi, T. et al. Direct observation of motor protein stepping in living cells using MINFLUX. Science 379, 1010–1015 (2023). DOI: 10.1126/science.ade2676
6 Alvelid, J. et al. 2022. Event-triggered STED imaging. Nature Methods 19:1268. DOI: 10.1038/s41592-022-01588-y
7 Willig, K. et al. 2022. Multi-label in vivo STED microscopy by parallelized switching of reversibly switchable fluorescent proteins. Cell Reports 35:109192. DOI: 10.1016/j.celrep.2021.109192