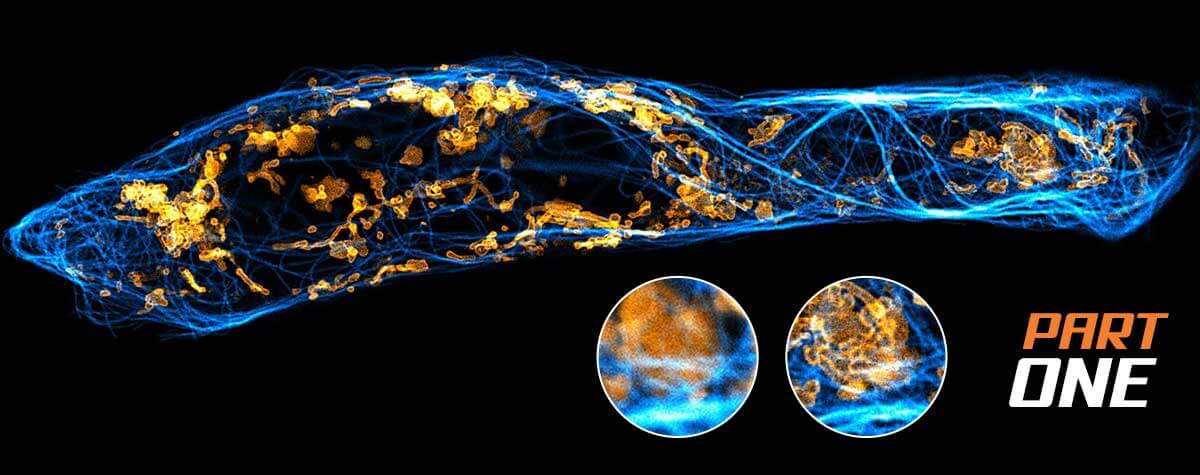
STED
Superresolution microscopy is a widely used research method for studying biological features beyond the diffraction limit, such as nuclear pores, clathrin coated vesicles, the periodicity of the actin skeleton in neurons, and viruses. In this article we will focus on the advantages of STED microscopy and its contributions to the virology field. We will provide insight into the advances in virus research made possible by STED microscopy and give a hint to were the journey might go.
and virology
Virus particles vary in size across different virus species. One of the smallest viruses, the parvovirus, has a particle size of approx. 20 nm while the largest known virus particles of the Mimivirus family possess a capsid size of up to 500 nm, which already compares to the size of a bacterium (Fig. 1). Therefore, due to their size, virus particles and their substructures can be studied in detail using super-resolution microscopy methods, such as stimulated emission depletion (STED) microscopy or single-molecule localization methods, which offer resolution capabilities of about 20 nm. Numerous studies using super resolution imaging techniques have already expanded the scientific knowledge of the Human Immunodeficiency Virus (HIV-1) 1, 2, Influenza A virus 3, 4, Herpes Simplex Virus (HSV) 5, Respiratory Syncytial Virus (RSV) 6, Vaccinia Virus 7, Adeno-Associated Virus (AAV) 8, Nipah Virus (Niv) 9, Adenovirus 10 and many more.
STED microscopy for virus research
The gold standard technique for imaging virus particles was and still is electron microscopy (EM). The main advantage of EM is undoubtable the Ångstrom-range resolution it provides, which exceeds the resolution capabilities of all other available imaging techniques. The extremely good resolution, however, comes at the cost of highly fixed specimens with low contrast. Furthermore, labeling and detection of specific proteins inside large assemblies and cells is challenging with EM. On the other hand, light microscopy methods, and especially confocal fluorescence microscopy, generate high-contrast images, with specific labels for the proteins of interest. However, the resolution of these techniques is limited to about half the wavelength of light (~200 nm). The Nobel Prize winning methods of superresolution microscopy, specifically STED microscopy and single molecule localization microscopy techniques, overcome this resolution barrier by exploiting the on-off switching of fluorescent dyes (find out more about how the donut changed the world). These techniques improve confocal resolution by roughly a factor of 10, achieving resolution capabilities closer to those offered by EM (see Fig. 1). Furthermore, the nanoscale resolution of STED microscopy combined with the less destructive sample preparation and labeling methods of fluorescence make it possible to study the structural information of native (unfixed) virus particles in the 100 nm-range 2. An example of how STED microscopy has contributed to HIV-1 research is detailed in the box on page 3.
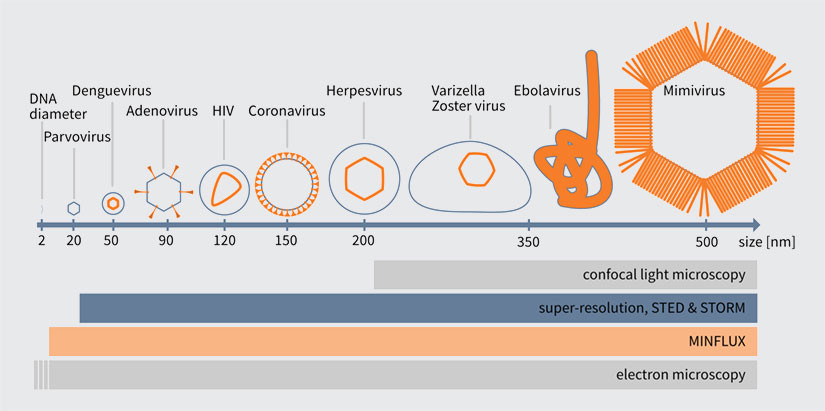
Figure1. Comparison of different virus particle sizes with the resolution of different imaging techniques.
Protein-specific labeling coupled with the high contrast imaging of light microscopy enables colocalization studies of different proteins to shed light on, for example, virus-host interactions. In conventional microscopy, however, our understanding of the protein position is blurred by diffraction. Superresolution microscopy techniques overcome this blurring effect and thus lend a much higher confidence to the results of colocalization experiments. STED microscopy is especially well-suited for these studies since resolution can be improved not only laterally, but axially as well, giving rise to isotropic resolution of up to 60 nm without any post-processing. Additionally, if two spectrally different dyes are used which can both be depleted by the same STED laser doughnut (e.g. abberior STAR ORANGE and abberior STAR RED, or Alexa594 and ATTO647N), there is no chromatic shift between the two channels. Because the central zero of the STED doughnut defines the point from which photons are emitted, if two channels share the same depletion doughnut, colocalization is intrinsically given. Various studies have used STED microscopy to visualize the HSV genome inside infected cells 11 or to study the virus-host interaction of the Influenza A virus 3, 4.

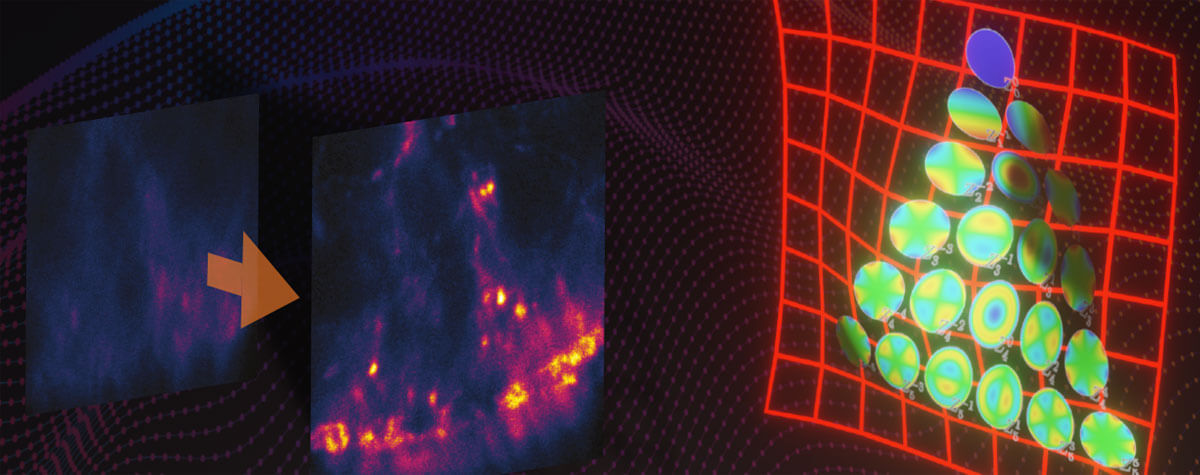
Figure 2. Env protein distribution (red/yellow) on single HIV-1 particles (green). Vpr.eGFP was used as a marker for HIV-1 particles. Env clusters on the virus particle surface were labeled by Fab fragments coupled to abberior STAR 635P. Only with super-resolution microscopy can single Env clusters be revealed (compare middle column (STED) and right column (confocal)). Images by Jakub Chojnacki (used with permission).
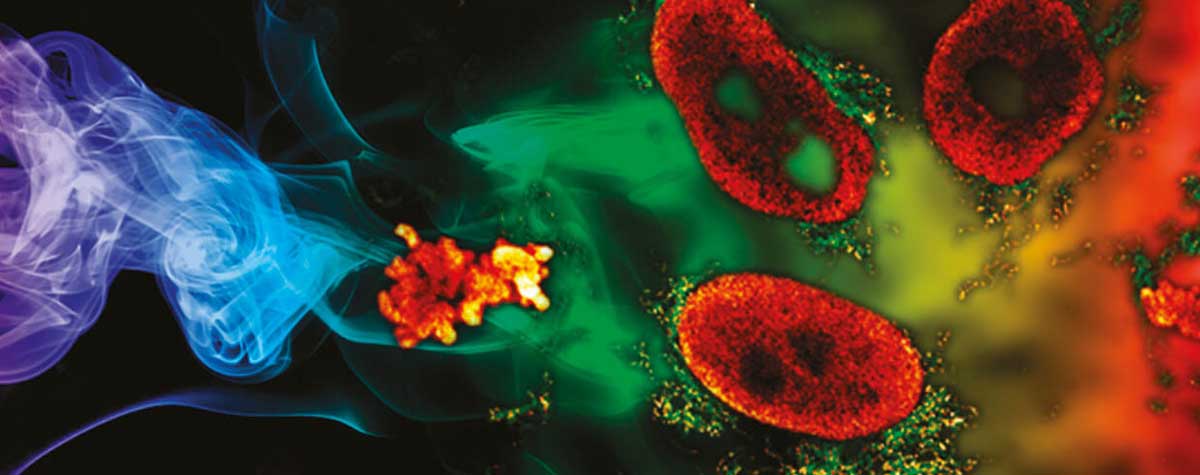
The possibility of live-cell imaging and the tracking of dynamic events over time is a great advantage of fluorescence microscopy, and STED microscopy uniquely combines this advantage with nanoscale resolution. Live-cell STED microscopy has proven valuable in many studies 12-14. A major concern in live-cell STED microscopy is bleaching and phototoxicity. While bleaching can be overcome by using photostable dyes or exchangeable fluorophores 18, minimizing phototoxicity is less straightforward. Nevertheless, with STED microscopy, it is often overlooked how resolution and laser power are interdependent. Due to the square-root dependence of resolution on STED power, high STED laser power is only needed to squeeze out the last few nanometers of resolution. In STED microscopy, the achieved resolution is tunable via the STED laser power and this can be increased only as much as required to answer a given scientific question. Furthermore, novel technologies such as pulsed STED combined with Adaptive Illumination methods (e.g. MINFIELD, DyMIN and RESCue) 15-17, allow for prolonged imaging at high resolution with low light levels and thus less phototoxic effects. An example on HIV-1 imaging is shown in Fig. 3. Here, it is also important to note that since a virus particle contains only a small number of proteins and hence labels, gentle adaptive illumination methods help minimize photobleaching, thus allowing one to image with higher resolution, or for a longer time in a live experiment.
Especially sparse events are difficult to image in EM and can usually only be found by correlative imaging of light microscopy and EM. Identification of sparse events, such as cell infection or virus particles binding to enter a cell, or nucleus, are the perfect target for STED microscopy. Since every STED microscope is inherently a confocal microscope (with the STED laser turned off), it is possible to scan the sample in confocal mode with a low magnification objective lens to easily identify rare events. After finding the event, a region of interest can be chosen to target the event, and the STED laser can be turned on to acquire a super-resolution image. The imaging of capsid positive objects after HIV infection of cells close to the nuclear pore 19 is one example of how STED microscopy can be used to study rare events.
Scientific research involving virus imaging requires a large range of imaging scales, from a single virus particle on the coverslip 2, to whole infected cells 19 or even intact living mice 21 to, for example, observe the immune response in lymph nodes. Thus, the ideal superresolution microscope needs to be flexible to perform all given imaging tasks. STED microscopes reliably meet this need and can be adapted to a variety of imaging experiments. Objective lenses and mounting media can be chosen as needed for the experiment. RAYSHAPE aberration correction allows STED and confocal imaging deep inside the sample by using a deformable mirror to correct for aberrations induced by the sample. With these tools at hand, the STED microscope is able to image across scales and highly adapts to the experimentalist’s needs.
A glimpse into HIV-1 studied by STED microscopy
Human Immunodeficiency Virus 1 (HIV-1) maturation occurs concomitantly with cleavage from the plasma membrane of its host cell. During maturation, the protease is active, and cleaves the polyprotein Gag into its components, leading to a rearrangement of the virus proteins. Mature HIV particles enter cells more efficiently. However, there is no apparent change of the surface protein (envelope protein, Env) structure or composition. Compared to other virus particles, only a few envelop proteins are incorporated on the HIV surface (7-14 Env proteins per virus particle). The question remains of how the internal morphological change influences the Env protein on the particle surface, and what the surface effect is that increases entry efficiency into new host cells.
In 2012, the research team of Hans-Georg Kräusslich answered these questions using STED microscopy, when the method was still in its infancy. In a collaboration with Stefan Hell’s group, they showed that Env proteins on single virus particles form clusters after virus maturation, and that this clustering is induced by maturation through an interaction between the Env tail and the internal virus proteins. Since virus particles (diameter ~120-150 nm) are much smaller than the diffraction limit of light, only direct observation by super-resolution microscopy was able to resolve this clustering (see Fig. 2) 1.
In a follow-up study at the University of Oxford, researchers were able to determine the mobility of the Env trimers on a single virus particle. This study showed that the HIV-1 lipid envelope is a low mobility environment due to its high lipid order. The standard technique to image mobility is Fluorescence Correlation Spectroscopy (FCS). However, since the virus particles are smaller than a conventional focused spot, intensity fluctuations due to particle movement can only be observed by a combination of STED and FCS, to reduce the focal volume 23.
The high lipid order observed led to a collaboration between labs in Spain, France, Germany and Australia to study the lipid environment of budding HIV particles in cells by STED-FCS. The study showed that around an HIV-1 budding site, the virus seems to generate its own lipid environment, which is likely due to the recruitment of specific lipids through the main structural polyprotein Gag 24.
In summary, major contributions to basic research on HIV-1 were achieved using STED microscopy. Also in the future, STED and super-resolution microscopy in general will be instrumental in the research on small structures such as viruses.
Conclusion and Outlook
In this article, we provide a brief overview of superresolution microscopy, focusing on STED microscopy and its contributions to virus imaging and virus research. Developments in STED microscopy instrumentation are rapid and, while the technique has already seen widespread use in scientific research, we are confident that it’s potential can only increase in the future. One should not forget that the first ever images on a home-built STED microscope were acquired only less than two decades ago. Since then, tremendous amounts of engineering and innovation have made STED microscopy what it is today: a reliable, easy-touse super resolution imaging technique that can be operated as intuitively as a confocal microscope, by everyone.
The potential of superresolution microscopy extends far beyond STED. The novel technique MINFLUX 22 offers isotropic resolutions up to 2 nm – the size of a single fluorescent molecule. Already STED microscopy was able to change the way we look at virus particles, so the impact MINFLUX will have on scientific research cannot be underestimated. This new level of resolution is expected to have a tremendous impact on virology research. MINFLUX tracking can be performed at extremely high speeds with true molecular precision, making it possible to observe rearrangements inside a single protein over time, or to follow single virus particles over a long time with unprecedented precision and speed.
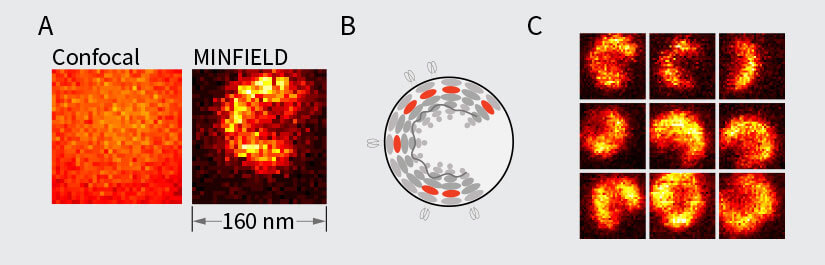
Figure 3. MINFIELD STED presented on human immunodeficiency virus type 1 (HIV-1 labeled with SNAP-tag between matrix and capsid in the Gag protein and stained with Siliconrhodamine). (A) Confocal (left) and MINFIELD STED (right) images with a field size of 160 nm. (B) Scheme of the immature HIV-1 particle with labels indicated in red. (C) Imaging examples of single HIV-1 particles.
1 Maturation-Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy, Chojnacki, J. et. al, SCIENCE, 26 OCT 2012: 524-528
2 Stimulated Emission Depletion Nanoscopy Reveals Time-Course of Human Immunodeficiency Virus Proteolytic Maturation,
Hanne, J., et al, ACS Nano 2016 10 (9), 8215-8222, DOI:10.1021/ acsnano.6b03850
3 IFITM3 Clusters on Virus Containing Endosomes and Lysosomes Early in the Influenza A Infection of Human Airway Epithelial Cells, S. Kummer et al., Viruses 2019, 11(6), 548
4 Visualization of early influenza A virus trafficking in human dendritic cells using STED microscopy, F. Baharom et al., PLoS ONE 12(6): e0177920.
5 Structural analysis of herpes simplex virus by optical super-resolution imaging, Laine, R., et al., Nat Commun 6, 5980 (2015). https://doi.org/10.1038/ncomms6980
6 Multicolor Stimulated Emission Depletion (STED) Microscopy to Generate High-resolution Images of Respiratory Syncytial Virus Particles and Infected Cells, M. Mehedi et al., Bio Protoc. 2017 Sep 5; 7(17): e2543.
7 Gray, Robert & Albrecht, David. (2019). Super-resolution Microscopy of Vaccinia Virus Particles. 10.1007/978-1-4939-9593-6_16.
8 Super-resolution imaging of nuclear import of adeno-associated virus in live cells, Kelich, Joseph M et al., Molecular Therapy – Methods & Clinical Development, Volume 2, 15047 [9] A stochastic assembly model for Nipah virus revealed by super-resolution microscopy, Liu, Q. et al., Nat Commun 9, 3050 (2018). https://doi.org/10.1038/s41467-018-05480-2
10 Tracking Viral Genomes in Host Cells at Single-Molecule Resolution, Wang, I-Hsuan et al., Cell Host & Microbe, Volume 14, Issue 4, 468 – 480
11 Visualizing the replicating HSV-1 virus using STED super-resolution microscopy. Li, Z. et al., Virol J 13, 65 (2016). https://doi. org/10.1186/s12985-016-0521-7
12 Live-cell imaging of dendritic spines by STED microscopy, Nägerl, V. U. et al., Proceedings of the National Academy of Sciences Dec 2008, 105 (48), 18982-18987; DOI: 10.1073/pnas.0810028105
13 Live-Cell Superresolution Imaging by Pulsed STED Two-Photon Excitation Microscopy, Takasaki, K. T. et al., Biophysical Journal, Volume 104, Issue 4, 770 – 777
14 Two-colour live-cell nanoscale imaging of intracellular targets, Bottanelli, F. et al., Nat Commun 7, 10778 (2016). https://doi. org/10.1038/ncomms10778
15 Adaptive-illumination STED nanoscopy, Heine, J. et al, Proceedings of the National Academy of Sciences Sep 2017, 114 (37) 9797-9802; DOI: 10.1073/pnas.1708304114
16 MINFIELD STED microscopy, Göttfert, F. et al, Proceedings of the National Academy of Sciences Feb 2017, 114 (9) 2125-2130; DOI: 10.1073/pnas.1621495114
17 Far-field optical nanoscopy with reduced number of state transition cycles, Staudt, T. et al., Opt. Express 19, 5644-5657 (2011)
18 Whole-Cell, 3D, and Multicolor STED Imaging with Exchangeable Fluorophores, Spahn C. et al., Nano Letters 2019 19 (1), 500- 505, DOI: 10.1021/acs.nanolett.8b04385
19 Analysis of CA Content and CPSF6 Dependence of Early HIV-1 Replication Complexes in SupT1-R5 Cells, Zila, V. et al., mBio Nov 2019, 10 (6) e02501-19; DOI: 10.1128/mBio.02501-19
20 Resolution scaling in STED microscopy, Harke, B. et al., Opt. Express 16, 4154-4162 (2008)
21 In vivo mouse and live cell STED microscopy of neuronal actin plasticity using far-red emitting fluorescent proteins. Wegner, W. et al., Sci Rep 7, 11781 (2017). https://doi.org/10.1038/s41598-017-11827-4
22 Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes, Balzarotti, F. et al., SCIENCE, 10 FEB 2017: 606-612
23 Envelope glycoprotein mobility on HIV-1 particles depends on the virus maturation state. Chojnacki, J., et al., Nat Commun 8, 545 (2017). https://doi.org/10.1038/s41467-017-00515-6
24 HIV-1 Gag specifically restricts PI(4,5)P2 and cholesterol mobility in living cells creating a nanodomain platform for virus assembly. Favard, C., et al., Science advances, 5(10), 2019, eaaw8651. https://doi.org/10.1126/sciadv.aaw8651