abberior news
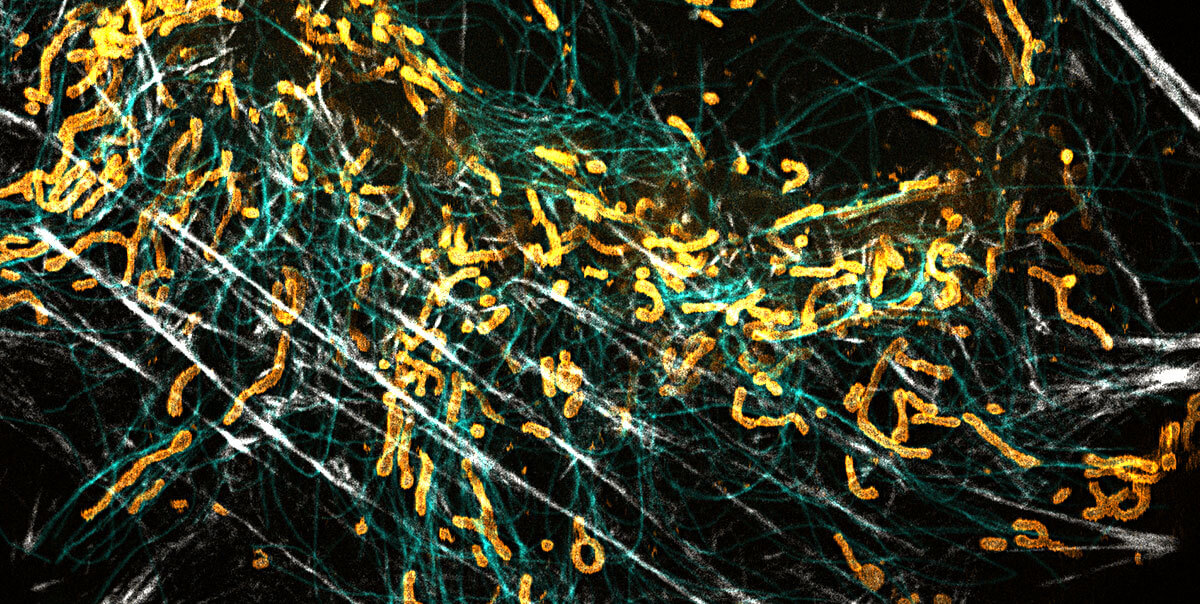
superhot superresolution
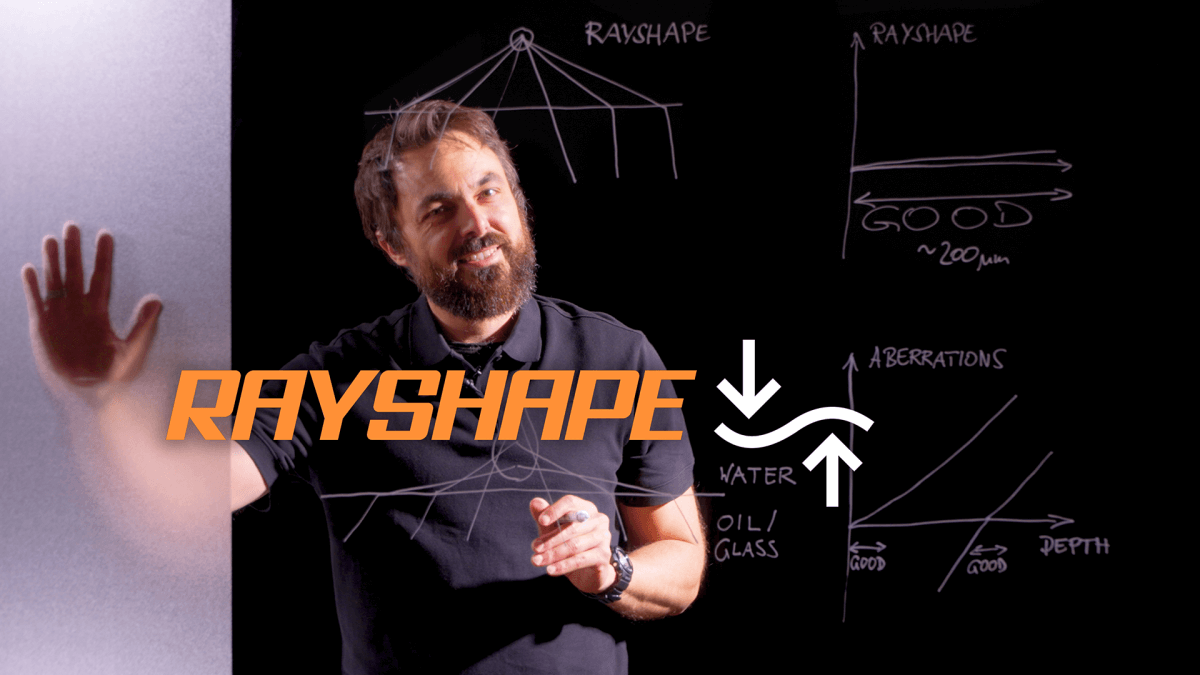
Watch Recording: Webinar on RAYSHAPE dynamic aberration correction
Hello RAYSHAPE! Bye-bye, aberrations!
As a confocal or superresolution microscopist you are most likely familiar with the frustration of blurred images and weak signals that are caused by aberrations, in particular when focusing deep into samples. Maybe you already tried correction collar objectives to tackle aberrations and are discontent with their performance.
In our webinar you will learn how RAYSHAPE uses a deformable mirror to dynamically eliminate aberrations even deep in the sample.
We are happy to welcome Jan Maximilian Janssen, group leader in the lab of Maren Engelhardt at the Institute of Anatomy and Cell Biology at JKU Linz, Austria, as guest speaker. He will show you how adaptive optics with RAYSHAPE helps him to take aberration-free images in thick human brain tissue to study axonal plasticity and the composition of specialized nano domains along the axon initial segment.
Maximilian’s talk will be complemented by the introduction of our application specialist Bastian Klußmann-Fricke into the principle and the capabilities of RAYSHAPE.
Watch recording of the webinar from June 20, 2024.
We at abberior love donuts
Stefan Hell overcame Abbe’s diffraction limit in light microscopy with the help of a donut-shaped laser beam. Since then, the donut has been abberior’s best friend. In STED, it de-excites fluorophores and increases resolution, providing us with exciting superresolution images. Even when resolution took another quantum leap with MINFLUX, we could rely on our good old friend, the donut, again. It simply changed jobs. Now it excites fluorophores and enables us to achieve unmatched spatial and temporal resolution with just a view photons. That’s why looking through a donut makes us smile! And they’re yummy too, of course!
Want to know more about our donuts? Read on:
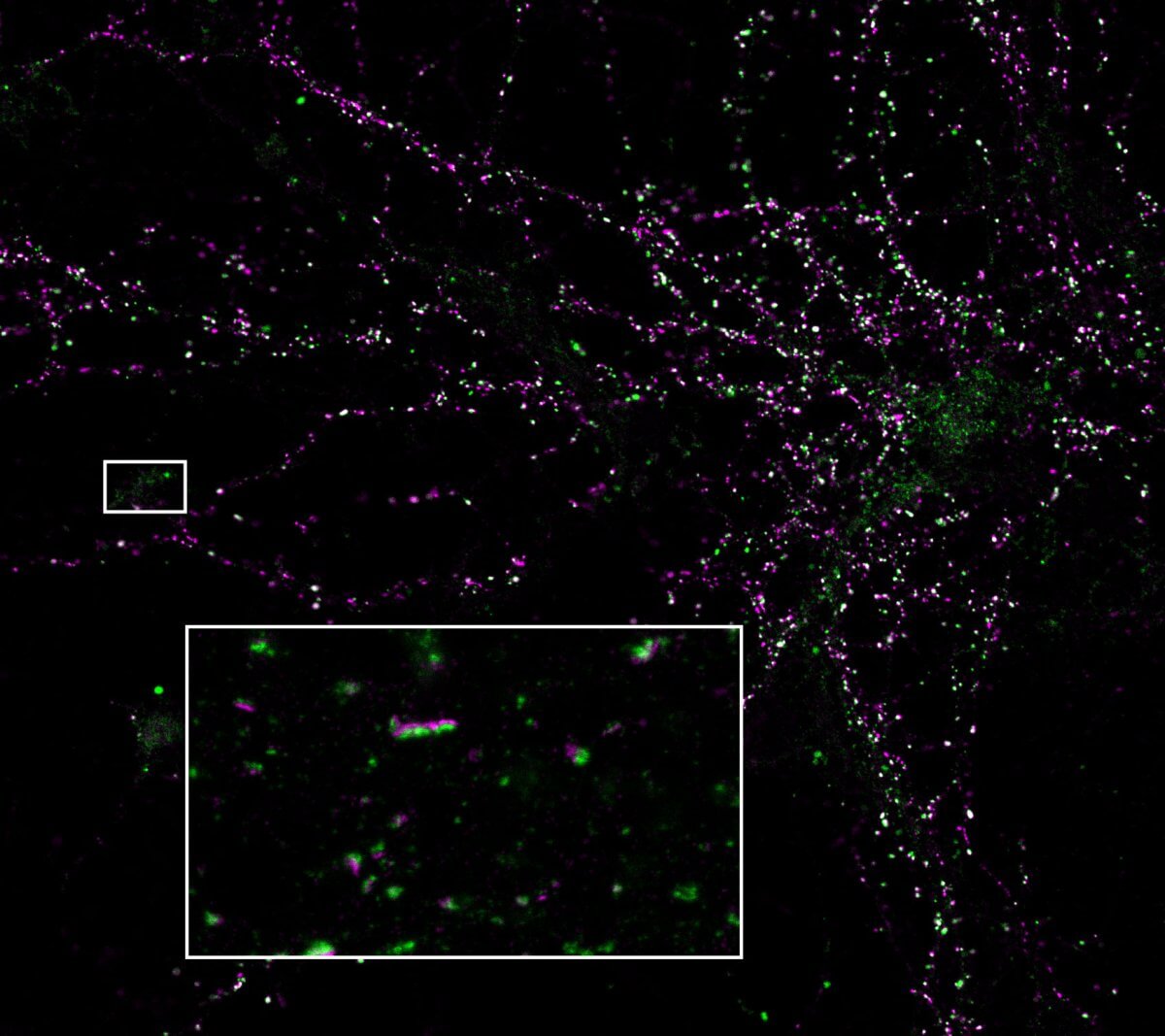
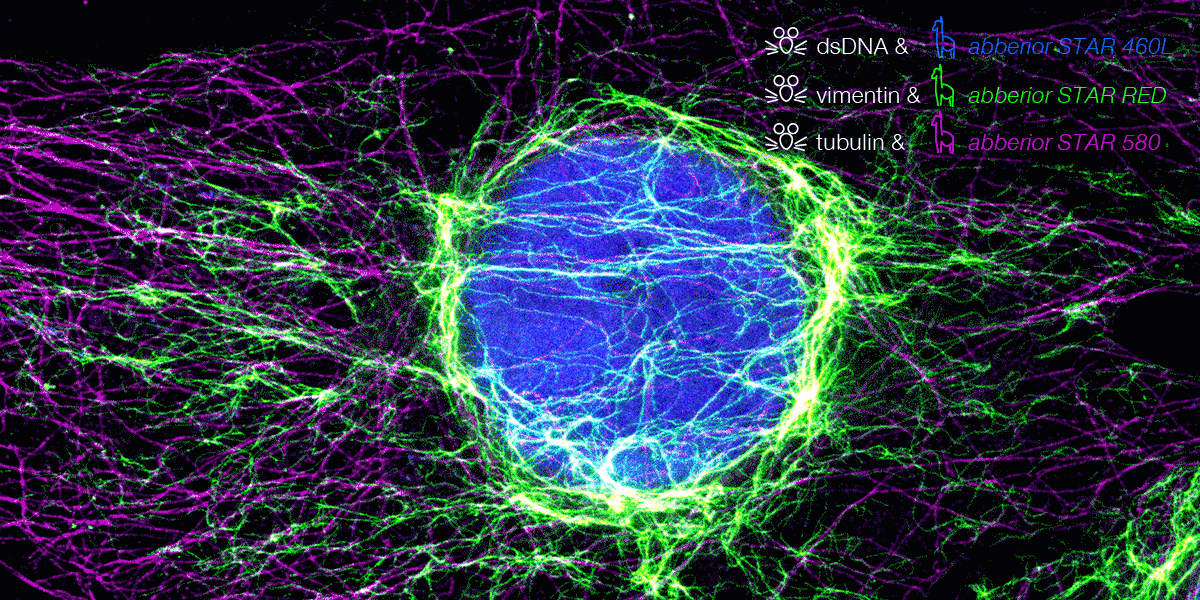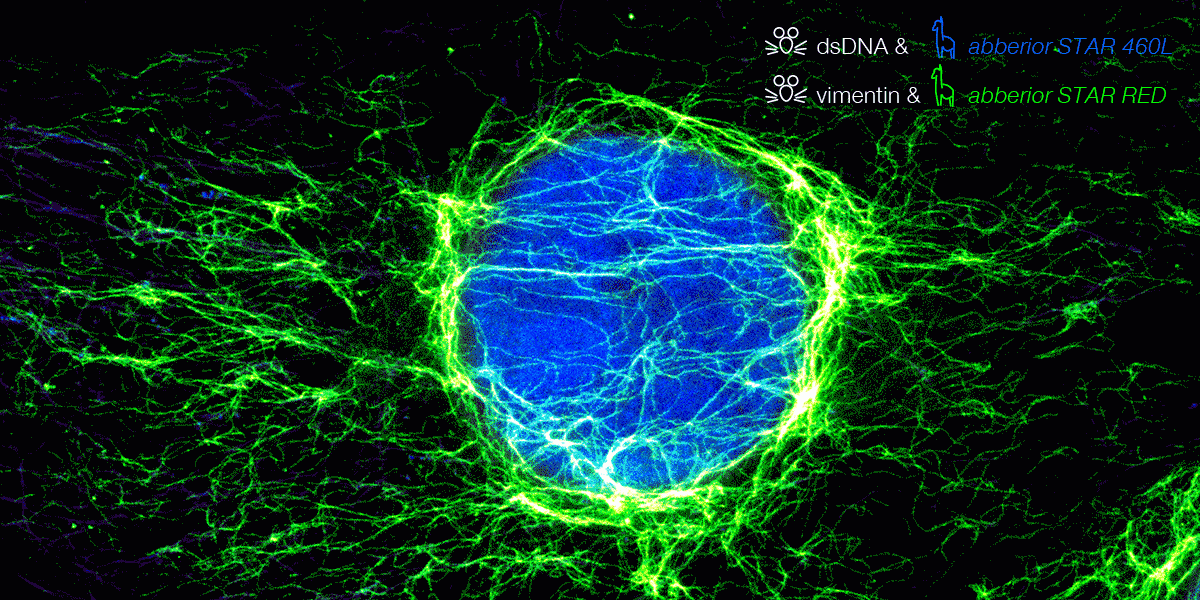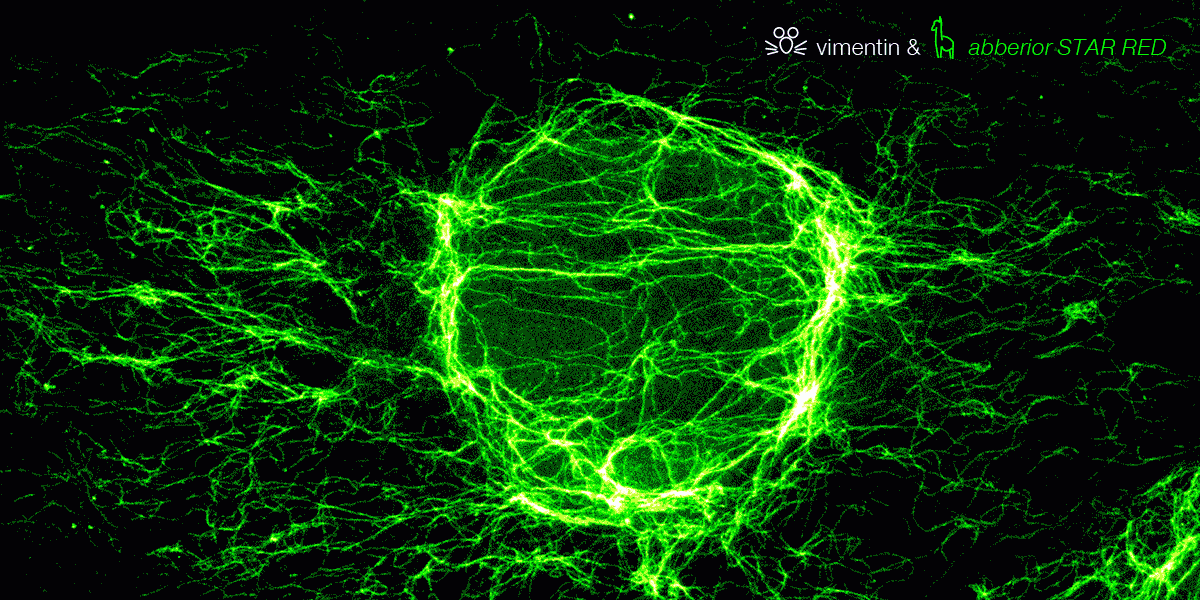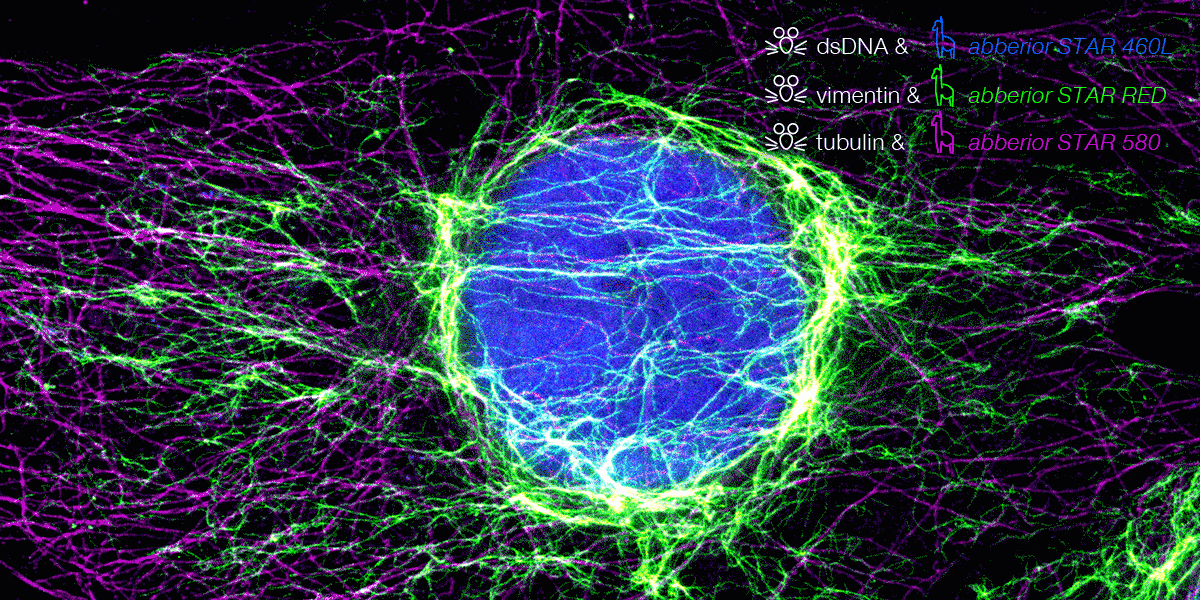
We are excited to announce our expanded partnership with Synaptic Systems, now offering primary antibodies coupled with abberior STAR dyes. These dyes are optimized for STED microscopy, providing precise neuronal protein detection with strong signals and low background fluorescence.
Discover how this innovation can elevate your research Read more >
Indirect immunofluorescence staining is a pivotal method for biomolecule labeling, yet its efficacy can be hindered by large complexes that form between primary and secondary antibodies and fluorophores. This causes large linkage errors, and artifacts due to the distance between the label and the target protein, therefore solutions which increase the proximity between the fluorophore and the target molecule are crucial. We present a groundbreaking solution by combining abberior FLUX dyes with Jackson ImmunoResearch’s AffiniPure-VHH™, polyclonal VHH fragment secondary antibodies.
Specifically optimized for single molecule localization techniques such as MINFLUX and STORM microscopy, abberior FLUX dyes ensure precise localization. Paired with JIR’s polyclonal VHH fragment antibodies (nanobodies*) which bind to multiple sites of the primary antibody, they ensure remarkable signal amplification, resulting in captivating and informative images. This innovative combination heralds a new era in immunofluorescence labeling, offering superior performance, full flexibility, and enhanced visualization capabilities.
Interview

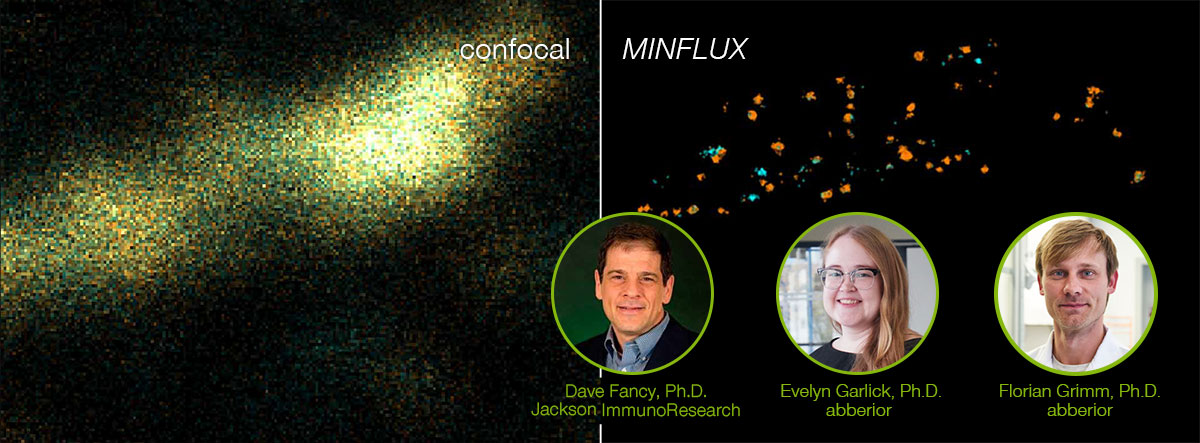
Dave Fancy, Ph.D., Jackson ImmunoResearch
Dave is the Chief Operating Officer for Jackson ImmunoResearch Laboratories. After gaining his Ph.D. from the University of Texas, Dave has specialized in the development of novel antibody conjugates of dyes, metal chelates, enzymes and DNA for use in the research. He has extensive experience in developing immunological products and services, with a focus on commercialization for the biotech research, diagnostic communities.

Evelyn Garlick, Ph.D., abberior Instruments
Evelyn comes from the UK and completed her Ph.D. at the Universities of Birmingham and Nottingham in 2022. Since then, she has been a part of the MINFLUX applications team at abberior Instruments, working to help users get the most out of their MINFLUX microscopes.

Florian Grimm, Ph.D., abberior dyes & labels
Florian, with a background in biochemistry and chemistry, was abberior’s first employee. With over a decade of experience with abberior dyes, labels, and microscopes, he now serves as a Business Developer. Florian is dedicated to finding innovative probes and maintaining key business relationships, driving abberior’s mission forward.
How does the format of AffiniPure VHH™ secondary antibodies help?
Fancy: The novel format of the VHH fragment is a 10th of the size of a conventional Ig antibody, this is good news if you want precise labeling where the fluorophore is close to your target protein because the primary/secondary complex is considerably reduced in size.
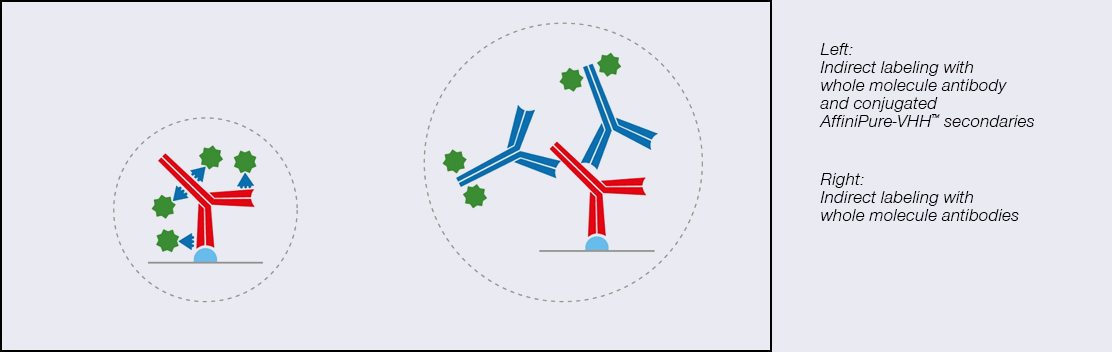
Figure 1. VHH fragments in comparison to conventional lg antibodies
I’ve used monoclonal nanobodies, why should I use a polyclonal one?
Fancy: The limitations of monoclonal antibodies are apparent when you need to achieve maximum brightness. The signal amplification achieved with polyclonal AffiniPure- VHH™ antibodies comes from the heterogenous population of antibodies, with different paratopes able to bind at many sites across the primary antibody ensuring it is maximally decorated with signal-producing fluors. Figure 1 demonstrates the advantages of polyclonal VHH antibodies, binding across the whole of the primary antibody, rather than being limited to a specific site/region/sequence, enabling higher labeling efficiency and brighter signal than the monoclonal format.
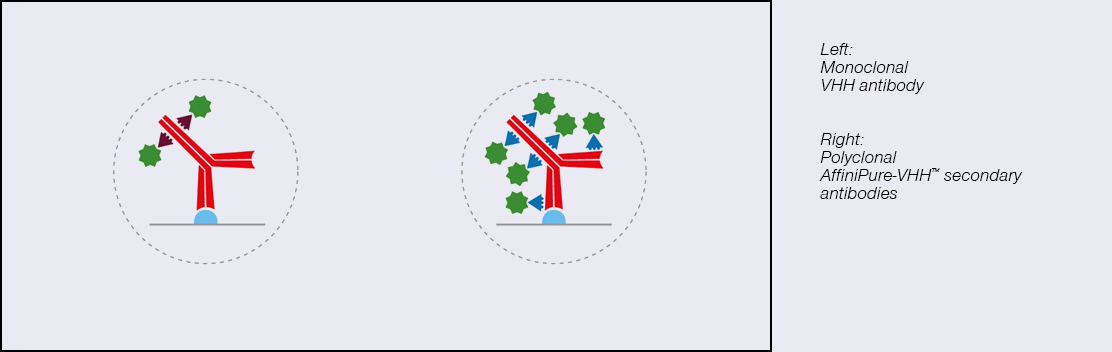
Figure 2. The polyclonal format of AffiniPure-VHH™ ensures maximum signal generated without compromising the signal density/resolution.
I’ve got multiple targets; will these work together?
Fancy: JIR AffiniPure-VHH™ antibodies are cross-adsorbed against commonly used species to enhance specificity and thus reduce off-target labeling, they can be used in combination to generate exquisitely specific multiple labeling images.
Access to an entire spectrum of dyes – shouldn’t this be phrased as a question?
Fancy: Indirect labeling using conjugated AffiniPure-VHH™ Secondaries gives researchers access to a greater range of fluorophores, with AffiniPure VHH™ Secondaries available conjugated to fluors from ultraviolet to far-red researchers can pick the right characteristics for their experiments which is useful for those identifying multiple target proteins or seeking dyes suitable for SRM and SM techniques.
What is MINFLUX microscopy?
Garlick: MINFLUX (MINimal photon FLUXes) is a single molecule localization technique that finds fluorophores using an excitation minima rather than maxima. This difference is what makes MINFLUX the most precise and photon-efficient way of localizing fluorescent molecules, capable of achieving 1 – 3 nm localization precision in all three dimensions. The unparalleled precision afforded by MINFLUX nanoscopy has many applications, giving impressive insights into biological structures at the nanoscale.
What are FLUX dyes?
Grimm: FLUX dyes are organic fluorescent molecules that have been developed and tested for MINFLUX microscopy. The main feature of this particular class of dyes is that they start to blink through the use of a special buffer system. This blinking feature enables our dyes to oscillate between an ON and OFF state, a crucial aspect as it ensures that not all dyes are active simultaneously, allowing for precise localization of individual dye molecules over time.
But the versatility of abberior FLUX dyes doesn’t end there. They’re also compatible with other single-molecule localization microscopy techniques such as STORM, opening doors to many research possibilities.
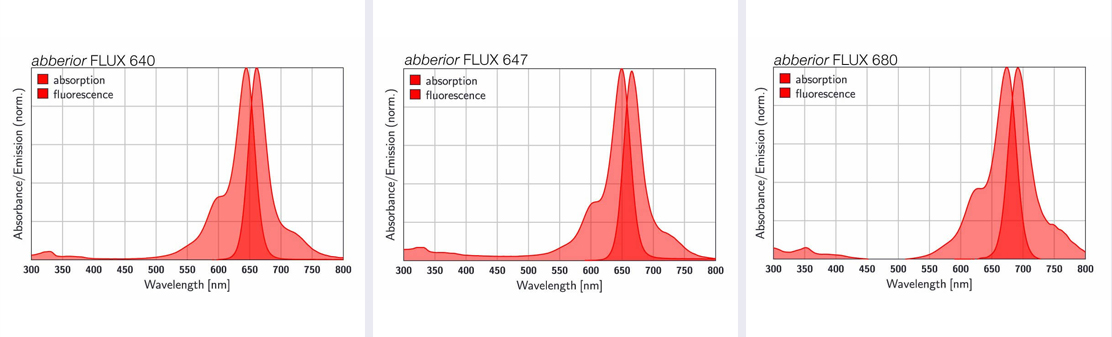
Figure 3. Absorption and fluorescence spectra of abberior FLUX dyes.
What is the advantage of FLUX dyes conjugated to AffiniPure-VHH™ antibodies?
Garlick: Dave explained that when using traditional primary and secondary antibodies, the distance from your epitope of interest to the fluorophore on your secondary can be ~ 20 nm. This is called linkage error. If you have a technique like MINFLUX that is capable of localizing a fluorophore with a precision of < 3 nm, reducing this linkage error is essential to better understand the arrangement of the epitope. AffiniPure-VHH™ Secondary antibodies can improve the linkage error, being much smaller than conventional Ig, they bring the fluorophore closer to the epitope, whilst keeping the flexibility of a traditional immunostaining approach.
How to use abberior FLUX dyes conjugated to JIR AffiniPure-VHH Fragment antibodies?
Garlick: FLUX-conjugated VHH fragment secondary antibodies can easily be incorporated into existing staining routines in place of your conventional secondary antibody. Detailed information can be found here: Protocols >
What is important when preparing a MINFLUX sample?
Garlick: Some special attention to your sample can help you achieve great quality MINFLUX results. In general, steps should be taken during labeling to limit background from both autofluorescence and unspecific labeling or binding. Make sure you incorporate a robust blocking and washing routine.
What dye to choose for single-color and which pair for two-color MINFLUX imaging?
Garlick: Our go-to dye recommendation for single-color imaging is FLUX 647. The best pair for two-color MINFLUX is FLUX 640 and FLUX 680 – these fluorophores are nicely spectrally separated in their emission and perform well in the same blinking buffer conditions, allowing simultaneous imaging of the two labels with the same excitation line.
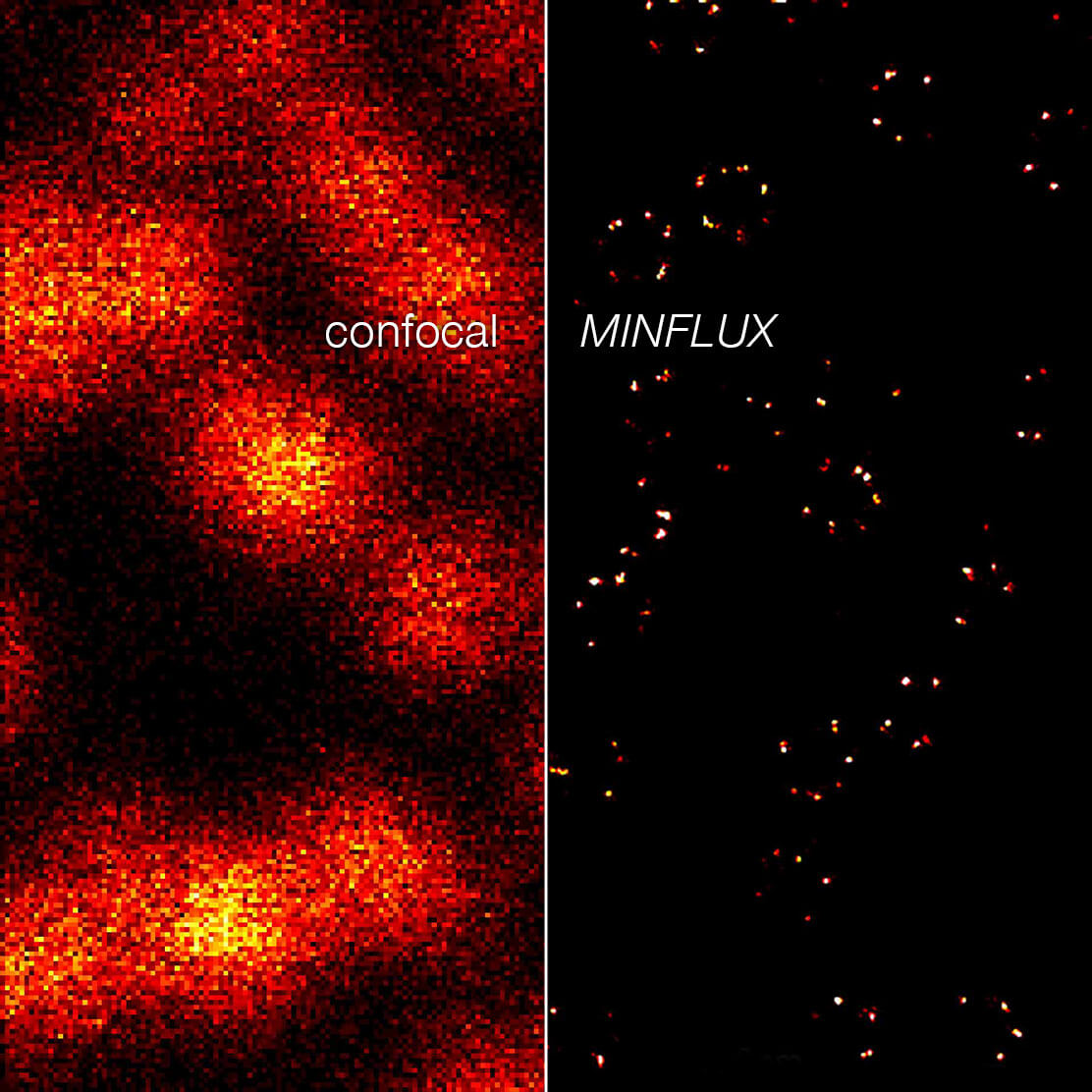
Figure 4. 2D MINFLUX nanoscopy of the nuclear pore complex subunits, labeled with abberior FLUX 647 conjugated to JIR AffiniPure-VHH Fragment antibodies (secondary nanobodies). In contrast to confocal microscopy, 2D MINFLUX allows visualization of the shape and arrangement of individual nuclear pore complex subunits. Here, we reach localization precisions of ~ 2 nm in raw localization data.

Figure 5. Two-color 3D MINFLUX revealing an inner and outer mitochondrial membrane marker. Cultured mammalian cells labeled with indirect immunofluorescence using JIR AffiniPure-VHH Fragment antibodies (secondary nanobodies) coupled to abberior FLUX 640 (orange) and FLUX 680 (cyan). MINFLUX enables the visualization and separation of both structures.
By now we can call it a tradition: We again took part in the Company Cup at the Göttinger Altstadtlauf on June 18, 2024. Spectators cheered on the runners, and after the race (at the very latest), everyone was in a great mood! Already looking forward to next year…!
Discover the revolutionary potential of live-cell fluorescence microscopy with our latest innovation – abberior Halo labels!
Traditional live dyes often face limitations due to the need for specific reactive groups for each target. However, the emergence of genetically encoded self-labeling protein tags like the HaloTag® eliminates this constraint, offering unprecedented flexibility for live-cell applications. Our HaloTag® labels ensure precise targeting with minimal off-target effects, whether you’re conducting live-cell imaging or examining fixed specimens. These labels are optimized for STED and confocal microscopy and guarantee unparalleled clarity and resolution.
Choose from six vibrant colors! Whether you’re studying live-cell interactions, tracking protein dynamics, or exploring intracellular structures, abberior LIVE 550, 590, and RED Halo labels offer endless possibilities. Explore further with our abberior STAR GREEN, ORANGE, and RED Halo labels, designed for cell impermeant labeling of HaloTag® fusion proteins. Whether on the cell surface of living cells or intracellular targets of fixed cells, these labels expand the horizons of your research.
Experience the future of live-cell imaging with abberior HaloTag® labels – where flexibility meets precision, and discovery knows no bounds.
HaloTag® is a registered trademark of Promega Corporation
100x sharper than a confocal microscope, 100x faster tracking than a camera – that is MINFLUX. abberior’s unique system reaches unprecedented spatio–temporal resolution in light microscopy and has already been used to address research questions in various fields since its introduction in 2016. Don’t miss the chance to learn what MINFLUX is capable of!
We are honored to welcome EMBL group leader Jonas Ries, his PostDoc Takahiro Deguchi and the Master student Christopher Heidebrecht as guest speakers, who talk about how they have taken advantage of MINFLUX’ unique abilities to investigate the structural dynamics of the motor protein kinesin in living cells.
Our abberior application specialist Clara-Marie Gürth complements Jonas’ presentation with an introduction to molecular tracking with MINFLUX.
Watch the recording of the webinar from January 17, 2024.
Twelve years after abberior’s foundation we are finally there: We moved into our brand-new headquarters in Hans-Adolf-Krebs Weg 6 at the North Campus of the University of Göttingen!
“The move reflects our strong growth since the company’s foundation,” says abberior CEO Dr. Gerald Donnert. ” Our team now comprises more than 130 colleagues – many from different scientific fields – and new colleagues are joining us every month. The new building offers ideal conditions for further developing our high-tech instruments, testing them together with customers and thus creating ever more powerful tools for the life sciences. We are very grateful that the City of Göttingen, the GWG, and the University of Göttingen have worked with us so excellently, making it possible to construct the building within a sporty timeframe.”
Read the press release here.
Light microscopy without aberrations that eat up your signal and blur the image – sounds like a faraway dream to you?
Well, it’s time for your dream to come true! Our RAYSHAPE aberration correction uses a deformable mirror to put aberrated rays back on track, restoring signal and focus even deep in the sample. And the best thing is that RAYSHAPE’s correction is dynamic: It continuously adjusts the deformable mirror as the focus moves through the sample, for zero aberration and a crystal-clear image from top to bottom.
Deep imaging has never felt better than with RAYSHAPE!
Explore everything that RAYSHAPE can do for your imaging here.
Live-cell fluorescence microscopy suffers from the limited number of commercially available live dyes as each target requires its specific reactive group. The recent development of genetically encoded self-labeling protein tags such as the HaloTag® overcomes this limitation and offers more flexibility to live-cell applications.
However, the covalent, irreversible attachment of the HaloTag® labels to the target protein favor photobleaching and exclude it from imaging applications that require temporary binding.
We now overcome these limitations: Here comes our exchangeable fluorescent abberior LIVE HaloX®! It is engineered to interact with the HaloTag® protein only transiently, allowing free exchange with fresh labels. In confocal and STED microscopy, this transient binding ensures that proteins can be detected in the living cell over extended periods as bleaching is no longer an issue. And hey, our HaloX® labels are combinable with the current HaloTag® system, so just get the new labels and go ahead!
Learn more about HaloX® in our webinar and watch the recording >
HaloTag® is a registered trademark of Promega Corporation
HaloX® is a registered trademark of Spirochrome AG. This product is covered by one or more license from Spirochrome AG and is intended for Research Use Only (RUO)
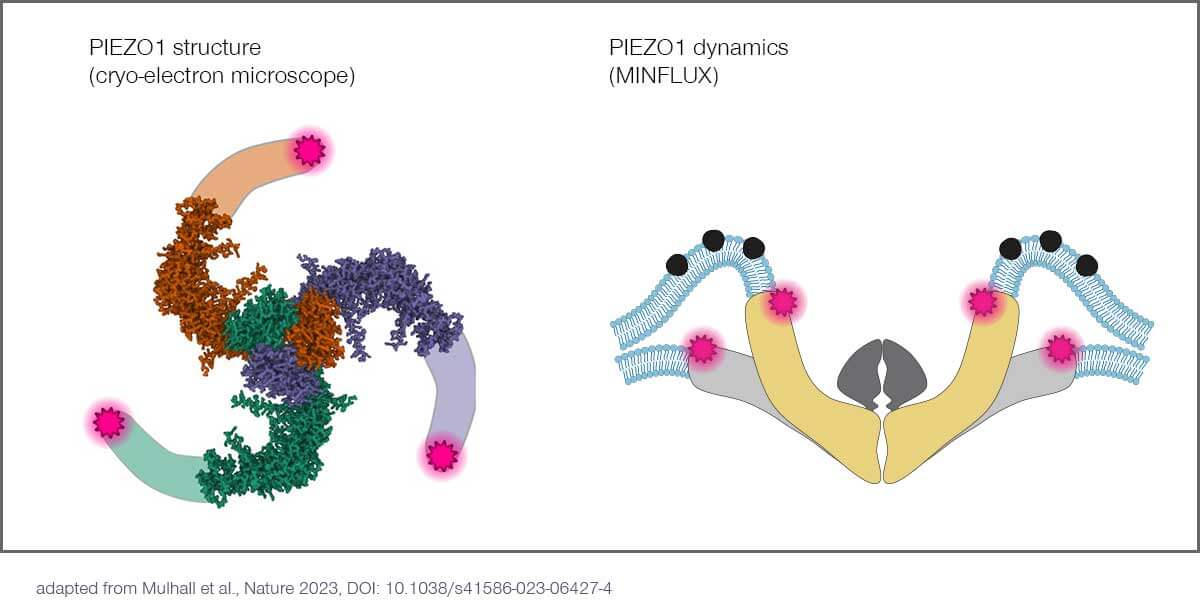
In the current issue of Nature, 2021 Nobel laureate and neuroscience professor Ardem Patapoutian, PhD, and his team at Scripps Research (La Jolla, US), unravel the conformational changes, with single nanometer resolution, of PIEZO1 ion channels in response to mechanical stimuli.
PIEZOs are a family of ion channels that adapt their shape and permeability in response to mechanical stimuli. PIEZOs consist of three identical protein subunits that form a central pore with three blades of transmembrane domains extending outwards and upwards, resembling a propeller. It has been proposed that these blades sense changes in membrane tension and gate the channel in response. Patapoutian received the 2021 Nobel Prize in Physiology or Medicine for his PIEZO discoveries.
Classical methods reach limits
Structural models of PIEZO1, developed via cryo-electron microscopy, have provided molecular details about the pore and the proximal part of its blades, but these models are incomplete and lack the distal one-third of the blades. Furthermore, cryo-electron microscopy – and related techniques such as crystallography and nuclear magnetic resonance spectroscopy – all require the protein of interest to be highly purified, and therefore struggle to provide contextual information about how proteins interact with their native physiological environment. These techniques also rely on extensive averaging to assemble molecular models due to their low signal-to-noise ratio and often fail to resolve distinct conformational states of individual proteins.
For PIEZO1, this means that the exact conformation of its blades, how they perceive mechanical force, and subsequently influence channel activity are unknown.
In the new study, first author and Scripps Research postdoctoral fellow, Eric Mulhall, PhD, and team applied the cutting-edge microscopy technique MINFLUX to tackle these questions. MINFLUX is a super-resolution light microscopy technique first published in 2016 by 2014 Chemistry Nobel laureate Stefan Hell. It offers unprecedented spatial and temporal resolution – on the order of ~1 nanometer and ~100 microseconds, respectively – even in living cells, making it possible to localize and track individual molecules, in their native state, in real time and three dimensions.
Monitoring PIEZO1 changing conformation with MINFLUX
The researchers combined MINFLUX with novel algorithms to investigate individual PIEZO1 molecules in cells with single nanometer resolution. Using a commercial MINFLUX microscope manufactured by abberior, they monitored how the blades of individual PIEZO1 molecules changed confirmation in their physiological context. By fluorescently labeling each blade at its most distal position – the propeller’s tips, so to speak – they were able to identify their exact position with nanometer precision and measure the distance between the blades. This in turn provided information about the protein’s conformational state. The scientists found that the blades are significantly expanded at rest due to the bending force exerted by the plasma membrane and that their conformational flexibility increased with distance to the central channel.
MINFLUX inventor Hell says: “This outstanding study from Ardem Patapoutian’s lab is exactly what I always dreamed MINFLUX would end up enabling: making groundbreaking physiological discoveries.”
As the measurements were performed directly in cells, the researchers were also able to observe the effects of a physiological stimulus. When exerting a hypo-osmotic shock, they witnessed how the blades stretched in response to an increased membrane tension and correlated this movement with channel activity.
To directly observe how PIEZO1 changes conformation and reacts to physiological changes in a living cell would not have been possible without MINFLUX.
Mulhall emphasizes: “This work provides a foundation for understanding how PIEZO1 is activated in a cellular context, and for the structural analysis of membrane proteins in their native environment.”
References:
- Mulhall et al.: Direct Observation of the Conformational States of PIEZO1. Nature 2023, DOI: 10.1038/s41586-023-06427-4
- Balzarotti et al.: Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 2016, DOI: 10.1126/science.aak9913
- Schmidt et al.: MINFLUX nanometer-scale 3D imaging and microsecondrange tracking on a common fluorescence microscope. Nat Commun 12 (2021), DOI: 10.1038/s41467-021-21652-z
Click here for the press release in english.
Click here for the press release in german.
Check out our latest articles about immunofluorescence labeling with antibodies and nanobodies.
“Why do superresolution microscopists love alpacas?”
It is a very simple yet very important fact: the localization precision of any microscope can only be as good as the size of the label allows.
Read article >
“Let the cells shine with immunofluorescence labeling!”
The most versatile and therefore most common strategy to bring the dye to the sample is immunofluorescence (IF). Learn how IF labeling works!
Read article >
Nanobody conjugated abberior STAR dyes
Nanobody protocol
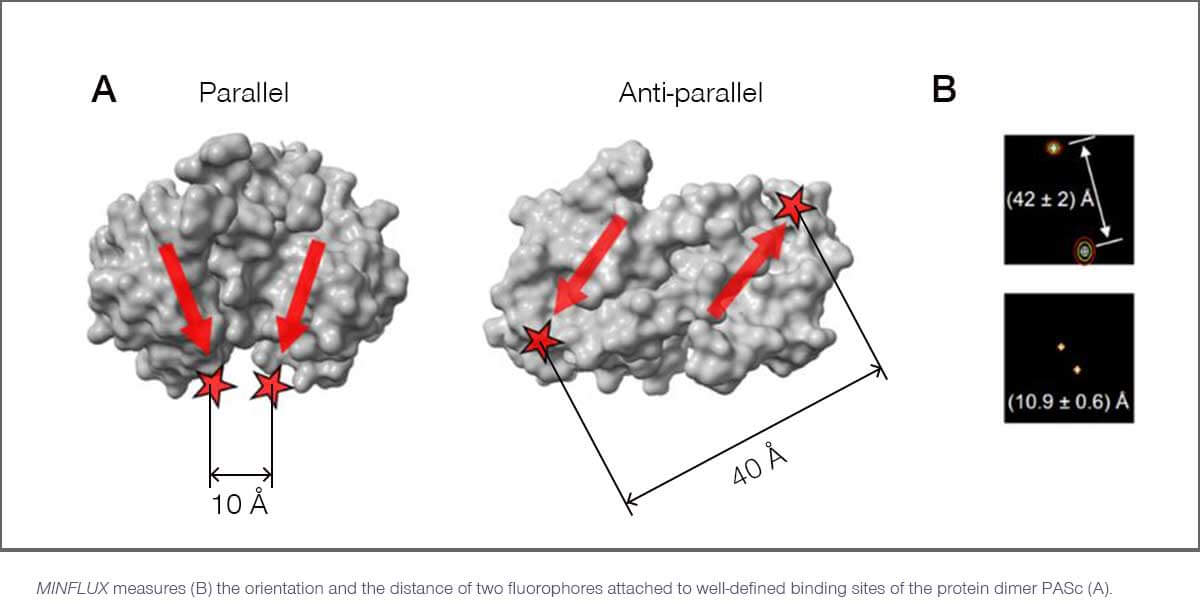
Researchers in Göttingen, Heidelberg and Berlin used a MINFLUX instrument from abberior to monitor the relative position of two fluorophores simultaneously, with sub-nanometer precision. Since these fluorophores were attached to well-defined sites on small proteins and oligomers, this provides direct access to the confirmation and orientation of these macromolecules.
In one experiment, Hell and colleagues took a dimeric protein called PASc. From crystallographic analyses, it is known that PASc exists in both parallel and anti-parallel configurations (see figure). This leads to different distances of the attached fluorophores, which MINFLUX can precisely quantify and discern with Ångström precision.
MINFLUX has a number of advantages over other methods when it comes to measuring distances between one and a few hundred nanometers. For example, FRET (Förster Resonance Energy Transfer) has a much more limited range. Also, unlike MINFLUX, it depends on the orientation of the fluorophores, and it cannot capture direction and orientation. Other techniques, like crystallography and electron microscopy, often rely on averaging a huge number of measurements, failing to provide information about individual molecules. But most importantly, they are not compatible with living cells.
In contrast, MINFLUX can directly observe individual cellular machines and their dynamics. It can directly measure distances, orientation and conformation with sub-nanometer precision and e.g. shed light on PASc and its structural rearrangement during the transmembrane signaling process with minimal invasiveness.
Steffen J. Sahl, et al.: “Direct optical measurement of intra-molecular distances down to the Ångström scale”, bioRxiv (2023)
Details >
Translation of the article “Was nie ein Mensch zuvor gesehen hat” in LaborJournal
JOURNAL CLUB
What no human being has ever seen before
The developers of super-resolution fluorescence microscopy were awarded the 2014 Nobel Prize in Chemistry by the Royal Swedish Academy. With MINFLUX, prize winner Stefan Hell has since improved its resolving power by another order of magnitude. Even conformational changes of individual proteins can now be tracked with nanometer precision on the millisecond scale. In this interview, the biophysicist explains how.
By Andrea Pitzschke
Have you ever watched proteins at work? No, not photometrically in an enzymatic assay, but in real life? Neither a conventional nor a confocal microscope will do for this. Because of the diffraction of light, the resolving power of normal light microscopes has physically predetermined limits of 200 to 300 nanometers. The size of proteins, however, is in the low nanometer range. Green fluorescent protein (GFP) with its 238 amino acids and 28 kilodaltons, for example, measures only 4.2 nanometers by 2.4 nanometers.
When additionally several molecules lie together, their contours become blurred. Making them visible in the midst of uninteresting molecules is the key advantage of fluorescence microscopy. But again, the informational value is limited: In small spaces, such as within the cell nucleus, even fluorophore-labeled molecules appear only as a luminous cloud. This reveals what a molecule’s favorite organelle is, but whether it is moving, interacting with other cell components, or just lying around lazily remains unknown.
This changed around the year 2000 when the group of Göttingen physicist Stefan Hell developed Stimulated Emission Depletion (STED) microscopy. STED initiated the era of super-resolution fluorescence microscopy, which also includes the methods described a few years later and known as Photoactivated Localization Microscopy (PALM) and Stochastic Optical Reconstruction Microscopy (STORM), respectively. With a resolution of 20 to 30 nanometers STED and PALM/STORM improved visible detail by a factor of ten, and their developers were awarded the 2014 Nobel Prize in Chemistry.
Underlying both techniques is the ability to selectively turn fluorescence signals on and off. PALM/STORM collects the signals of individual fluorophores and uses them to construct a microscopy image. STED, on the other hand, suppresses the fluorescence of molecules away from a focused center point by using a ring-shaped laser beam of intense light with a lower-energy wavelength around the center point . Both STED and PALM/STORM rely on collecting as many photons as possible from fluorophores. Meanwhile, time passes during which molecules move and fluorophores may bleach irreversibly.
Superresolution 2.0
Stefan Hell, of course, developed his trick with the ring-shaped “donut” laser beam further and finally presented Minimal Photon Fluxes (MINFLUX) with his research group in 2017 (Science. doi.org/f9kgvq). MINFLUX localizes fluorophore-labeled molecules, as well. Once again, a “donut” laser scans a two-dimensional sample for this purpose, but this time it searches for the weakest possible fluorescence signals. From the fluorescence data, the MINFLUX system iteratively determines in which direction and with which step size the excitation minimum of the laser must move to approach a fluorescent molecule until it finally detects a minimal fluorescence signal – in the best case, the background level. This is because the “donut” laser’s central excitation minimum then coincides precisely with the fluorophore. From the known position of the laser the position of the fluorescence molecule can be inferred.
The advantage is obvious: While PALM/STORM searches for signal maxima, MINFLUX spares fluorophores. Photobleaching is no longer an issue. Compared to PALM/STORM, a hundred times fewer photons are sufficient for the same localization accuracy. If the position of a fluorophore is already known with nanometer accuracy, even with only 20 to 40 photons MINFLUX manages to detect any further change in position with nanometer accuracy. This not only makes single-molecule tracking a hundred times faster than before, but also reduces the resolution limit by another factor of ten to the molecular scale of one to three nanometers.
No end in sight
Of course, the group of Stefan Hell, who is a director at the two Max Planck Institutes for Multidisciplinary Sciences in Göttingen and for Medical Research in Heidelberg, has not been idle since 2017. Their current MINFLUX version (Science. doi.org/j95s) uses three orthogonal pairs of beams instead of the “donut” laser to scan samples along all three spatial directions in increasingly fine increments. A look under its hood: With the help of phase and amplitude modulators, a 640-nanometer laser beam is shaped to produce a pair of beams with a defined phase difference. Their destructive interference produces an intensity pattern with a line-shaped rather than donut-shaped minimum in the focal plane. Two such beam pairs cover the x and y dimensions. For 3D images, a pair of beams in the z dimension is added. By changing the phase differences, all excitation minima can be shifted with an accuracy in the Ångström range.
What makes line-shaped excitation minima superior to the “donut” laser? The accuracy with which MINFLUX can localize a structure is determined by the steepness of the intensity transition at the boundary of the fluorescent molecule and background noise. If this boundary is abrupt and, in addition, the background intensity is low, the transition is steep. And this is precisely the advantage of the current MINFLUX microscope: Its line-shaped excitation minima inherently provide a higher slope compared to donut-based systems. As a result, the unexcited center of the laser is more precisely defined and fluorophores can be more sharply distinguished and localized.
The whole thing even works when the scanning lasers are tracking a moving fluorescent molecule. Accordingly, MINFLUX is suitable for molecule localization as well as for molecule tracking. Hell adds: “The imaging mode uses activatable dyes and delivers sharper images than PALM/STORM or STED. For molecule tracking, non-switchable fluorophores that emit more photons can be used instead.”
Gamechanger
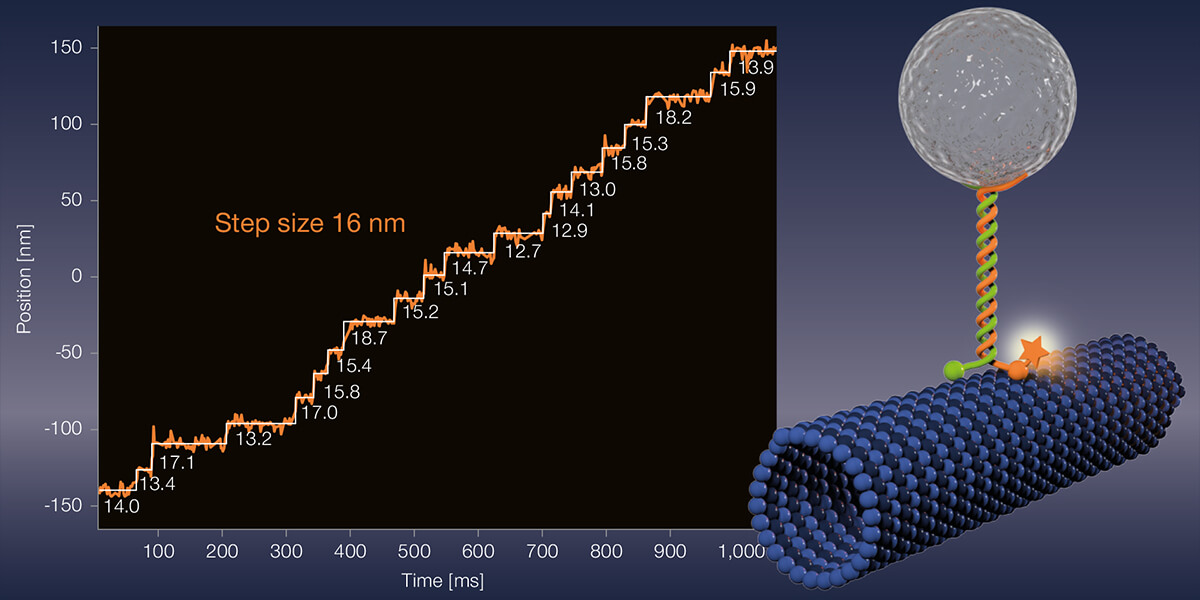
Meanwhile, Stefan Hell tersely dismisses the new gain in resolution: “Yes, our interferometric MINFLUX brings roughly another 30 percent improvement compared to the 2017 instrument.” But the main point of their current publication is to demonstrate MINFLUX’ application potential to the scientific community, says Hell. “The detail of the microscopy images is unparalleled. A gamechanger! For the first time, even conformational changes of proteins can be observed.” Hell’s words virbate with enthusiasm. Apparently, choosing a suitable demo object was almost a minor matter. “Let’s apply the method to something,” he told his co-workers. In the end, they chose the motor protein kinesin-1, which moves organelles, vesicles, and other cell components along microtubules inside the cell. Kinesin-1 exists as a dimer. Each monomer includes a head region binding microtubules and containing a catalytic domain, and a long stalk and tail portion interacting with the cargo to be transported. Hydrolysis of ATP changes the conformation of the head and neck regions, and the motor protein takes an eight-nanometer step.
The Hell group’s publication (Science. doi.org/j95s) appeared at the same time as an article by Jonas Ries’ group at EMBL in Heidelberg, Germany, who also studied kinesin-1 movement with MINFLUX (Science. doi.org/kbgn). “This was purely by chance, even though both groups are based in Heidelberg,” Hell recalls. “We didn’t realize the thematic overlap until the work was well advanced and we decided to submit the manuscripts to the same journal on the same day. We didn’t know each other’s specific content, but we have a good relationship, even though it wasn’t a collaboration.” While Hell’s group at the Heidelberg MPI studied the movement of the motor protein in vitro using their interferometric MINFLUX and established fluorescent dyes, Ries’ group at EMBL worked with live cells, the “donut” laser MINFLUX and an advanced fluorophore. The results of both projects confirm each other.
New scales
What exactly did Hell’s project involve? The experimentalists led by first authors Jan Otto Wolff and Lukas Scheiderer labeled different amino acid residues of kinesin-1 by maleimide coupling with the red-emitting fluorescent dye Atto647N. Since the fluorophore binds via cysteine residues, of which kinesin-1 has almost a dozen, it was necessary to mutate these or introduce artificial cysteine residues at selected positions. Finally, the Heidelberg researchers added fluorescently labeled kinesin-1 to microtubules immobilized on coverslips. As expected, kinesin-1 moved forward with eight-nanometer steps in the presence of ATP. This was hardly surprising.
What is unusual about the experiments of the Heidelberg scientists is that MINFLUX facilitated observations under physiological ATP concentrations in the range of one millimolar for the first time. Until now, the nucleotide had to be strongly diluted in order to throttle the running speed of motor proteins to be able to observe anything at all. But because MINFLUX requires so few photons, it can record fast movements. That puts the new record of spatiotemporal resolution at 1.7 nanometers per millisecond. “That’s 50 to 100 times faster than was previously possible,” Hell emphasizes.
Apart from speed records, the experiments also answered the long-standing question of when the motor protein binds and hydrolyzes ATP. In its 1HB state, kinesin-1 is bound to microtubules only with its leading head. In the 2HB state, the heads of both dimers interact with their respective microtubule-binding sites. Using a slowly hydrolyzable ATP analog (ATPγS), Hell’s group achieved a 36-fold slow-motion effect and found: ATP binds in the 1HB state when the unbound head is between the previous and next binding sites. ATP hydrolysis occurs only after the unbound head has moved to its next binding site.
Commercial MINFLUX devices are in use at EMBL as well as at research institutes in Jena, Shanghai, and Beijing. There are even two devices at the U.S. National Institutes of Health (NIH). “Publication numbers are skyrocketing. MINFLUX is one of the hottest microscopy techniques at the moment,” Stefan Hell explains winking. And he readily provided Laborjournal with further details.
Lab Journal: In 2021, your group published how a homebuilt fluorescence microscope can be upgraded to a MINFLUX instrument (Nat Commun. doi.org/gjnkp7). Would that also be possible with your new interferometry concept?
Stefan Hell In my Heidelberg lab, there is a self-built system that we use to explore physical limits. However, I advise users to purchase a new complete system. Observing the sample conventionally, for example via eyepieces, as is the case with a commercial system, makes sample handling and thus MINFLUX experiments easier.
In the 3D interferometry MINFLUX, pairs of lasers scan a sample in the x, y, and z directions. Does this happen simultaneously or sequentially? Do the two approaches work at different speeds?
Hell In our most recently published paper (Science. doi.org/j95s), we do it sequentially. This is not problematic because switching from one direction to the next takes place in fractions of a millisecond. In contrast, the commercial MINFLUX system localizes simultaneously along all three directions. There are good arguments for both approaches.
Is the autofluorescence of biological samples – also compared to conventional fluorescence microscopy – a problem? Is there anything that needs to be considered in particular during sample preparation?
Hell For samples, the general requirements of fluorescence microscopy apply. When you do imaging, you need photoactivatable or switchable organic fluorophores. When you want to track the position of an isolated molecule, the fluorophore does not need to be switchable. Any good fluorophore can be used, but fluorophores in the orange and red emission spectra are preferable because of the autofluorescence at blue and green excitation. There are no additional requirements.
Companies rarely mention the nanometer dimensions of their fluorescent dyes. Can and should this information be included by default?
Hell Fluorescent dyes are around one to two nanometers in “diameter”. It is important that a user is aware that a fluorescence microscope only images and tracks fluorophores. It cannot see molecules that do not fluoresce – this includes the biomolecule that a biologist is ultimately interested in. When you use a microscope with a resolution of one to five nanometers, it is therefore important to be aware of the distance between the fluorescent molecule and the biomolecule in order to draw the right biological conclusions.
Simplified, one could compare this to a bicycle tail light at night? Something is shining, but you don’t know where and how far you should swerve?
Hell Exactly. The light could come from a road bike just as well as from a mountain bike. You need to know how the taillights of a road bike differ from those of a mountain bike to deduce what you’re looking at. You never see the bikes themselves.
Does it then make any sense at all to develop even smaller fluorophores? After all, their linkers already introduce a certain size and thus blur. SNAP and HALO tags, for example, are already two to three nanometers long. Would it be better to look for linkers that are as rigid as possible, that do not wriggle and thus suggest that the linked protein is moving?
Hell Fluorophores are small enough. But one should indeed minimize the linkers. We used a maleimide linker with kinesin-1. It is much shorter than an antibody label, whose fluorophores can be a good five to ten nanometers away. Anyone using labeled antibodies must be aware that they are observing their target molecule from a certain distance.
The smallest fluorophores to date are about one nanometer in diameter. Can we make them any smaller?
Hell Hardly. A molecular structure that can fluoresce has a size of about one to two nanometers, which is due to the physics of the delocalized light electrons in the organic fluorophore.
So do you see a need for optimization more in the fluorescent dyes or in their linking technologies?
Hell Definitely in the latter. In the past, the focus was on fluorophores that were as super-bright as possible. Thanks to MINFLUX that is no longer necessarily an important criterion. But suddenly the spacing problem is prominent because of the high resolution.
With MINFLUX, in the observed region only a single molecule may carry a fluorescent label. Does one simply equimolarize the required dilution?
Hell In imaging, dilution happens by photoactivating and deactivating the fluorophores. Only one is turned on at a time, neighboring ones do not interfere. The activation beam provides the correct dosage. Commercial MINFLUX systems are also designed in such a way that the separation is integrated into the acquisition algorithm so that the user does not have to worry too much about it. In tracking mode, on the other hand, you have to determine the dilution empirically, so that in the oberserved region there is only exactly one molecule at a time.
Can a protein fold so unfavorably that it shields its fluorescent label? After all, MINFLUX measurements detect only a few photons in a small area …
Hell Yes, this cannot be ruled out. But the problem exists in general and is not typical for MINFLUX.
How many different positions of a protein should be fluorescently labeled for reliable localization?
Hell In principle, one position is sufficient. But for single molecule studies, you should label different positions to get meaningful statistics. That’s why my PhD students Otto Wolff and Lukas Scheiderer recorded thousands of kinesin-1 tracks to understand the biology of its movement.
You now know how kinesin-1 moves along microtubules. What other projects are you working on?
Hell Our study has shown what is possible today in terms of detecting protein dynamics and what you can get out of it. But the opportunities for biology are just opening up. I trust biologists to know best what questions can be answered on the millisecond scale with nanometer-precise microscopy. I look forward to every discovery that others will make with it. Perhaps a researcher still unknown today will win a Nobel Prize using it.
To what extent has your Nobel Prize influenced your career as a scientist? Certainly many things have changed for you, such as overflowing lectures and countless applications by PhD students?
Hell I can concentrate on science better than before and have a lot of time for my research group. That is very important to me. To be honest, I don’t give any lectures. I take the liberty of politely turning down bureaucracy, long lecture tours, and reviewing applications. I was lucky that I was quite young when I received the Nobel Prize. So I was able top it off with my people. When the Nobel Prize was awarded, the resolution limit was 20 nanometers. Now it’s one to two nanometers. That’s really fun.
Many thanks to LaborJournal and Andrea Pitzschke
Light microscopy is booming. Optical tricks make single proteins in cells visible and allow to observe complete organs. Now smart microscopes emerge which adjust to living tissue in no time – and watch the brain thinking.
„MINFLUX und MINSTED do not only see ten times smaller details, they also require a hundred times less fluorescence photons and are therefore a hundred times faster than PALM/STORM”
MIT Technology Review 5/2023
Read the full article in German here.
After last year’s successful restart, we again rocked the Company Cup at the Göttinger Altstadtlauf on July 5, 2023! The atmosphere in the city center was vibrant as always and the spectators’ support carried everyone to the finish line. We had lots of fun together – and see how a few sports-induced endorphins put a grin on everyone’s face…!
Almost a decade ago, the inventors of superresolution fluorescence microscopy were honored with the Nobel Prize for Chemistry by the Royal Swedish Academy. In the meantime, one of them, Stefan Hell, has stepped up resolution power by another order of magnitude. Now, even conformational changes of single proteins can be observed with nanometer precision on the scale of milliseconds. In an interview with German magazine LaborJournal, the biophysicist explains how.
“[With MINFLUX], photobleaching is not an issue anymore. Compared to PALM/STORM, 100 times less photons are sufficient for the same localization precision.”
LaborJournal 6/2023
Read the full article in German in LaborJournal
More information about MINFLUX
„MINFLUX is capable of tracking kinesin even amid the bustle of living cells.”
Powerful microscope captures motor proteins in unprecedented detail. Nature, June 8, 2023
“[with MINFLUX] … they are able to track kinesin very precisely. And when you have a live cell system, that’s even more spectacular.” Michelle Digman, University of California
Powerful microscope captures motor proteins in unprecedented detail. Nature, June 8, 2023
Read the full feature in Nature
More information about MINFLUX and “How the donut changed the world”
Selected references:
- Deguchi et al., Science 379, 1010 (2023)
- O. Wolff et al., Science 379, 1004 (2023)
- Balzarotti et al., Science 355, 606 (2017)
- Schmidt et al., Nat. Commun. 12, 1478 (2021)
Indirect immunofluorescence staining is one of the standard approaches for labeling biomolecules of interest, although these antibodies form huge complexes that cause some linkage error between the fluorophore and the biomolecule of interest. Therefore, it is essential to bring the fluorophore as close as possible to the molecule.
Here we go: Because they are ten times smaller – compared to conventional IgG antibodies – our new nanobody conjugated abberior STAR dyes offer much better access for superresolution microscopy. This significantly minimizes linkage errors compared to the standard IF staining method.
From now on, the combination of abberior STAR dyes conjugated with polyclonal secondary nanobodies (AffiniPure-VHH Secondaries) from Jackson ImmunoResearch offers a new superior solution for IF labeling. While the abberior STAR dyes provide optimal brightness and photostability in STED microscopy, JIR’s polyclonal nanobodies allow binding to multiple binding sites of the primary antibody. This provides high signal amplification and exciting image results are just a click away.
Get first-hand information and learn more about how our new nanobody conjugated abberior STAR dyes significantly improve superresolution imaging.
Watch the recording of our webinar on May 24, 2023, 16:00 (CEST) >
More information about our nanobody conjugated abberior STAR dyes:
Shop abberior STAR RED >
Shop abberior STAR 580 >
Shop abberior STAR 460L >
“But it (MINFLUX) was also well suited, for the first time, for tracking single fluorophores with simultaneous nanometer and millisecond precision.”
Physics Today 76 (5), 14 (2023)
Read the full article with Nobel Laureate Stefan W. Hell and Jonas Ries of the EMBL in Physics Today >
More information about MINFLUX and “How the donut changed the world!”
Translation of the article “Ein Potenzial, das noch in keiner Form gehoben ist” in the popular science journal Spektrum.de >
MICROSCOPY
A potential that has yet to be tapped into
In 2014, Stefan Hell shared the Nobel Prize in Chemistry for developing STED microscopy. Building on STED, he has now devised a revolutionary new fluorescence microscopy method.
By Verena Tang
People who use microscopes today can see details that were beyond their imagination just a few decades or even years ago. Stefan Hell, co-winner of the 2014 Nobel Prize in Chemistry, has made decisive contributions to this progress. Based on his Nobel-worthy discoveries he has devised a revolutionary new microscopy technique that observes fluorescent molecules with a spatial precision of less than one nanometer. Protein movements can thus be tracked with nanometer accuracy within milliseconds. He spoke with “Spektrum.de” about the ultimate limits of fluorescence microscopy and told us where he sees the greatest potential.
Spektrum.de: Professor Hell, until you invented STED microscopy about 30 years ago, there was good reason to believe that objects could only be resolved to about 200 nanometers. Now we have arrived at a few nanometers. Was there a point in the last 10, 20 years where you thought: This is the end – it doesn’t get any sharper than this?
Stefan Hell: I knew that it was conceptually possible to get down to molecular resolution, that is, a few nanometers. But there were reasons why we weren’t getting there. If you had asked me 10 years ago whether it would be possible one day, I would have answered: In principle, yes, but not with today’s methods and according to current knowledge. Nonetheless, I would have been optimistic that it would be possible at some point.
And so it came to pass …
Yes. Our new MINFLUX and MINSTED methods actually allow us to get down to molecular scales, that is, to separate molecules that are only molecular distances away from each other.
Let’s take a step back. How do you specifically make individual molecules glow?
All these methods, no matter what they are called – STED, PALM, or STORM (see “Beyond the diffraction limit”) work according to the same principle: on-off. If I want to separate and distinguish two molecules, I make sure that one of them cannot glow while the other does, and vice versa.
How exactly do you control that?
With STED, you control it with a light beam that says: there you are on and there you are off. So, I use laser beams to determine where “on” is and where “off” is. With this method, several molecules can also shine simultaneously in one place. In PALM/STORM, on the other hand, this is done by an intrinsic switching mechanism that stochastically turns individual molecules on and off. This works, for example, by using the thermal activation energy or by stochastically illuminating the sample. You get a fluorescence signal, and then you have to find out as precisely as possible where it comes from.
But?
In PALM/STORM, this is not done precisely enough. Therefore, we used elements of STED for that. This way, I can locate the glowing molecule with much greater precision. And in combination with the individual molecules turning on and off, you achieve very high resolution.
“The methods allow, for the first time, a robust and solid separation of molecules at the molecular scale”
So, you have brought two approaches together and thereby achieved molecular resolution.
We have combined the strengths of the two methods in a non-trivial way. That is, you end up separating single molecules, as with PALM/STORM, but you localize differently. To do that, you use a donut-shaped laser beam – or something similar, but a beam that has a point of zero intensity – and that allows you to determine the position much more precisely. By picking the strengths of the two methods and combining them, we came up with the new methods called MINFLUX or MINSTED. They allow, for the first time, a robust and solid separation of molecules at the molecular scale.
With MINSTED, you write, you can even resolve in the Ångström range …
No, it is not the resolution that is in the Ångström range, but the localization precision. That is, I can find out where the fluorescence molecule is with a precision of just a few Ångström. But that itself is one to two nanometers in size. Thus, it would be wrong to say that I have Ångström resolution – that’s impossible because I’m always separating molecules, and fluorescence molecules are at least one nanometer large. But it is quite useful to determine the position more precisely than the size of the molecule itself.
For example, to see to which specific location in a molecule a fluorophore binds?
Exactly. But let me just briefly provide context for the high resolution: With STED, you get down to 20 nanometers. That’s where the world of proteins begins. They are between 2 and 30 nanometers in size, and only now have we begun to cover that range with these new methods.
“We have now reached a scale where the size of the markers plays a role and where the limits of fluorescent labeling per se have a massive effect”
And yet, as you always need fluorescent markers, couldn’t you use them to elucidate molecular structures someday?
Yes, it has to be said that plainly: a fluorescence microscope sees fluorescent molecules. This is often misunderstood – but for good reason. In the past, the structures you looked at were much larger than the fluorescence molecule. Therefore, you didn’t have to worry about the fact that the fluorescence molecule itself has a dimension – that it was not identical to the protein you wanted to study. But of course, I never see the protein! I can’t see it at all because only the fluorescence marker glows. Simply put, this means that we have now reached a scale where the size of the marker plays a role and where the limits of fluorescent labeling per se have a massive effect.
What is the minimum size a molecule must have in order to fluoresce?
The physics of organic molecules requires molecules to be about one nanometer in size for the emission of visible light. And it is not only about the size of the fluorophore because it is connected to the protein via a linker. As such, the distance to the actual protein structure can be a couple of nanometers. That means you have a spatial mismatch between the fluorophore and the biomolecule you want to see. This problem used to be irrelevant when resolution was worse.
Allow me to make a casual comparison: it is exactly the same when you look at a beautiful painting from far away and up close, the Mona Lisa, for example. Seen from afar, she appears wonderful, but when you look into her eyes with a magnifying glass, you see the graininess, cracks, and brushstrokes of the painting.
Do you combine ultrahigh-resolution fluorescence microscopy with images from cryo-electron microscopy (cryo-EM) to elucidate structures? The methods complement each other, don’t they?
Yes, that’s what’s being done. You can correlate all these high-resolution methods. That certainly makes sense, but you have to keep one thing in mind: Cryo-EM images of biomolecule complexes are indeed high-resolution, beautiful. The only thing is: most of the time, those are averaged values. That’s because in cryo-EM, you’re looking at electron densities. But if I scatter the electrons coming from the electron microscope onto a protein, I get an immensely noisy image. So, experts take about 500 to 1000 images, superimpose them, average the values, and calculate the exact positions of the atoms. And here you see the limitations of cryo-EM: biological reality is usually not as nicely regular as cryo-EM usually suggests.
Imagine you had to explain to an alien what people on Earth look like. You could take 1000 pictures of women, average them, and say: this is a woman. But in reality, not a single one will look like that. When the alien walks down the street, he’ll say: wait a minute, the women all look different. It’s the same with the cryo-EM.
“We see some biological diversity that we recognize: They don’t all look the same in detail”
But isn’t that exactly where the technologies complement each other?
Absolutely, of course they complement each other. We see that, for example, in the nuclear pore complexes that we’ve been working on. When we look at those with fluorescence, we never, ever have in mind the accuracy and resolution of cryo-EM. There, resolution is actually in the Ångström range. But with fluorescence, we do see some biological diversity that we recognize: They don’t all look the same in detail.
What achievement would be the next personal highlight for you?
When you look at MINFLUX or MINSTED, the high resolution immediately jumps out at you. You’ve achieved that now; there’s probably nothing more to reach in terms of localizing the fluorescence molecules.
But there is something that is really exciting and in which I see huge potential: the dynamics of the molecules, that is, how they change their position in space. When the fluorophore is coupled to a protein, MINFLUX can detect how the fluorophore “wiggles” or moves better than any other technique. That is, when the fluorophore is attached to something, I can see the smallest deflection in space, the smallest movement in a short time, 100 times faster than with a camera. That’s a game-changer when it comes to capturing the motion or the conformational change of biomolecules tagged by the fluorophore.
What’s the problem with a camera?
To date, it works like this: You attach molecules to a protein, for example, to look at how the protein moves. Then you take a picture of the cell or parts of it with the camera and watch the movement of the dye. The dye leaves a diffraction spot of the emitted fluorescent light on the camera. This is also the case with PALM/STORM. Afterwards, you calculate the center – that is, the focal point – of the diffraction spot and say: this is where the dye was. With this procedure, I need a lot of fluorescence emissions per timepoint. Otherwise, I cannot calculate that focal point precisely. And that’s why the procedure is relatively slow – and it’s currently the only one that’s really well established.
Such techniques have been used, for example, to measure the steps of kinesin on microtubules. It was possible to see that its center of mass moves with eight-nanometer steps. To do this, however, the process had to be artificially slowed down so that only one step took place every third of a second. But proteins don’t move that slowly because otherwise, we wouldn’t be able to live. In other words, the ATP concentration was massively reduced because the localization process simply could not keep up otherwise.
To measure the dynamics directly is a prerogative of light microscopy
But now you have shown that it can also be done in real-time.
If I want to study movement in a living cell or in an artificial system, I can use MINFLUX to see the movement of fluorescent molecules attached to something 100 times faster – with the same accuracy. Or conversely, I accept the slowness but measure much more accurately. And then, the Ångström potentially plays a role because it can be quite important whether the fluorophore moves 8.2 or 8.4 nanometers. Then it’s not about resolution, but about how accurately I measure a movement or conformational change. Electron microscopy can’t do that because there the sample is frozen and in a vacuum. Measuring dynamics directly is a prerogative of light microscopy.
That means you’re just getting started?
We have a wide expanse ahead of us. It’s still very early, but in five to 10 years, MINFLUX could help in pharmaceutical development, for example, because it gives us the opportunity to follow dynamics precisely – for example, to study in detail how two molecules come together. I could label an active ingredient and measure how the dynamics of a protein change under the influence of this particular active ingredient. I see tremendous potential in pharmaceutical development, and it hasn’t been tapped into yet, in any way or form. People aren’t thinking that way yet.
Beyond the diffraction limit
To visualize structures in living organisms, biology works with fluorescent markers: dyes that bind to specific structures in a cell and glow when excited with light of a specific wavelength.
A natural limit restricts the resolution of such images: diffraction of light means that two objects closer than about half the wavelength of the emitted light can no longer be perceived separately. Because visible light has wavelengths between 400 and 700 nanometers, that limit is around 200 to 300 nanometers. Since the 1990s, scientists have been developing methods to circumvent this natural resolution limit. They all have one thing in common: by switching fluorescence on and off, they ensure that only one or a few molecules that are closer than 200 nm shine at a time.
STED
The technique developed by Stefan Hell is based on the prevention of fluorescence by stimulated emission depletion. As in classical microscopy, a laser beam excites fluorescent molecules in a specific area to glow. A second “donut-shaped” laser beam takes away the fluorescence ability of all molecules by stimulated emission – except those located in a very small area in the center of the “donut”. The smaller this area around the zero point of the STED laser, the fewer the molecules that can glow at the same time and the sharper the image. The sample is systematically scanned with this combination of on and off beams.
PALM/STORM
Here, the fluorescent molecules are initially off – that is, they are in a state where they cannot fluoresce when irradiated with excitation light. When switched on, the sample is first illuminated with turn-on light so weak that statistically at most one molecule is switched on randomly within the diffraction limit of 200 to 300 nanometers. The initially unknown position of the switched-on molecules is then determined on the basis of the diffraction pattern produced by the fluorescence of individual molecules on a camera. In this process, the maximum (or “centroid”) of the diffraction pattern is equated with the position of the molecule projected down onto the focal plane. This is called “localizing.” Then the molecules thus registered and localized are turned off and a new round of molecules is turned on.
This method, called PALM (“photo-activation localization microscopy”) or STORM (“stochastic optical reconstruction microscopy”), uses fluorescent molecules that are actively switched on and off or do so spontaneously (“blinking”). While the sequential switching on and off of individual molecules allows their separation, finding the center of their diffraction pattern reveals their position. Applying this to (almost) all molecules in the sample yields a sharply resolved image. The resolution is comparable to that of STED images.
MINFLUX
MINFLUX combines elements from the STED method and stochastic approaches. As in PALM/STORM, it uses molecules that can be repeatedly switched on and off. First, a maximum of one fluorescent molecule per diffraction region is statistically activated – that is, it is brought to a state where it can fluoresce under appropriate illumination with excitation light. Crucially, an excitation beam with an intensity zero in the center – i.e., again a donut-shaped laser as in STED, but this time for excitation – then scans the sample piece by piece. The position of the donut, and thus the donut zero, is controlled to within less than a nanometer, which is why it is always precisely known.
If a fluorophore is located exactly in the center of the excitation beam, the molecule remains dark because the intensity of the excitation beam is zero at that point. In this case, the position of the fluorescence molecule has been found exactly; it must be identical to the known position of the donut zero. If, however, the zero position is just next to the molecule, it glows faintly. Because the intensity of the excitation laser increases toward the donut ring, the number of emitted photons reveals the distance of the molecule from the zero point and thus the position of the molecule: the fainter it glows, the closer they are to one another. In the MINFLUX method, the donut-shaped excitation beam scans the sample piece by piece, optimally minimizing fluorescence per irradiated laser power. The better one succeeds in spatially overlapping the zero point with the molecule, the more precisely its location can be determined. In this way, the scientists obtain highly precise molecule positions. In combination with the sequential switching on and off of all fluorophores, the images achieve resolutions of 1 to 3 nanometers, that is, to the size of molecules.
MINSTED
MINSTED also combines the strengths of single-molecule microscopy with the precise localization of the STED method. Unlike MINFLUX, however, both a switch-on and a switch-off laser (STED) are used. Switchable fluorescence molecules are also used here. First, one molecule per diffraction limit is activated to switch on stochastically. The scanning of the sample works the same as with MINFLUX, but with a laser setup like STED – an excitation beam and a donut-shaped switch-off beam. Thus, the intensity of a fluorescent molecule is greatest when the center of the donut is directly above it. As the relationship between the distance from the zero point and the intensity is known, localization is based on this relationship. With MINSTED, it is now possible to localize fluorescing molecules to within a few Ångstroms, or tenths of a nanometer.
Many thanks to Spektrum and Verena Tang.
More information about MINFLUX and “How the donut changed the world!”
For the first time, the precise motion of the motor protein kinesin-1 as it walks on microtubules in living cells was measured by researchers from abberior, the European Molecular Biology Laboratory (EMBL), and Utrecht University.
Kinesin belongs to a class of motor proteins that transport large cargo to their required destinations in the cell. It can be pictured as a small molecule with two legs, which it uses to literally walk along microtubule-tracks inside the cell. Motor proteins are essential for many processes required for life, and consequently there is a huge interest in understanding their exact function and dynamics.
In the past however, the dynamics of kinesin on microtubules could not be studied under physiological conditions inside living cells. First, its individual steps are only a few nanometers long – far less than what conventional light microscopes can resolve. Second, they move so fast that no tracking technique could follow. The single molecule fluorescence tags used to label the motor proteins were simply too dim to provide reliable information on the spatial and temporal scales required here.
In a study now published in Science, Deguchi at al. used a superresolution microscopy technology called MINFLUX to investigate kinesin under physiological conditions. MINFLUX (MINimal photon FLUXes) was first published in 2017 by Nobel Laureate Stefan W. Hell and colleagues. It uses the limited photon budget of single fluorescent molecules much more efficiently than conventional techniques. In short, MINFLUX finds the exact position of a fluorescent molecule by actively aligning the central intensity null of a donut-shaped excitation beam onto it, until fluorescence emission is down to zero. In this case, every further photon that is not emitted is very useful, because it increases the confidence that the molecule is indeed located exactly at the position of zero intensity. The idea that photons that have never been emitted can carry positional information is the breakthrough concept behind MINFLUX. Once you wrap your head around it, it allows to localize and track single fluorescent molecules attached to e.g. kinesin, with unprecedented nanometer spatial precision and submillisecond temporal precision.
Using a commercial MINFLUX system provided by abberior, Deguchi at al. visualized the dynamics of a motor protein for the first time in living cells with a sufficiently small, non-disturbing label, yet a label that could be accurately tracked, thanks to MINFLUX. This allowed them to study motor proteins in near physiological conditions, i.e., in an environment that is much closer than before to what happens in living organisms. In the next stage, this paves the way for investigating how stepping behavior is altered by changes of kinesin’s natural environment, e.g. by the presence or absence of cargo or various microtubule-associated proteins, or under certain medical conditions.
Paul Selvin from the University of Illinois Urbana-Champaign, who published the first groundbreaking work on how kinesin walks on microtubules almost 20 years ago, says “This is a fantastic paper with beautiful data.” Christian Wurm, Head of Application at abberior adds: “The publication by Deguchi and colleagues is a prime example, where the unprecedented speed and accuracy of MINFLUX enabled researchers to make breakthrough discoveries. We expect this to be only one of many studies of this type.”
A similar study was concurrently published by Wolff et al., also in Science. Here, an optimized MINFLUX-setup with even higher precision was used. This enabled Wolff and colleagues, including personnel now at abberior, to observe smaller sub-steps and rotational movement of kinesin in vitro under physiological buffer conditions.
Ultimately, these findings are not limited to kinesin or motor proteins. MINFLUX opens the possibility to quantitatively record the precise structure and dynamics of molecular machinery in living cells, in general. This will provide significant insight into many processes that are essential to living systems, with the potential to better understand defects and diseases.
Selected references:
- Deguchi et al., Science 379, 1010 (2023)
- O. Wolff et al., Science 379, 1004 (2023)
- Balzarotti et al., Science 355, 606 (2017)
- Schmidt et al., Nat. Commun. 12, 1478 (2021)
Click here for the pressrelease
MINFLUX allows an everyday 3D resolution of 2 – 3 nm and offers about 100 times faster tracking than with a camera-based system.
Don’t miss this chance to listen to the inventor of STED and MINFLUX.
“If I want to study movement in a living cell or in an artificial system, MINFLUX allows me to see the movement of fluorescent molecules attached to something 100 times faster – with the same accuracy.”
Read the full interview with Nobel Laureate Stefan W. Hell in Spektrum >
More information about MINFLUX and “How the donut changed the world!”
abberior LIVE ORANGE mito is a new bright fluorescent probe that stains the structure of cristae in living mitochondria. Simply place the probe on your sample and immediately study the beauty of mitochondrial movement. Due to the special design of the probe, repeated excitation over a long period of time is possible without toxic effects or bleaching.
For beautiful STED images of mitochondria cristae in mammalian cells, please take a look at our sample gallery >
Are you interested in testing abberior LIVE ORANGE mito? Request a sample or order right away >
Order by December 31, 2022 and choose between three packages:
L abberior DYES DELUXE at a 15 % discount
Order a contingent worth € 3,000 and choose freely from our dye-labeled secondary antibodies on demand.
XL abberior DYES DELUXE at a 20 % discount
Order a contingent worth € 5,000 and choose freely from our dye-labeled secondary antibodies on demand.
XXL abberior DYES DELUXE at a 25 % discount
Order a contingent worth € 7,000 and choose freely from our dye-labeled secondary antibodies on demand.
Your package is valid until December 31, 2023 or until you reach your chosen package limit.
Has the Nobel Prize for Click Chemistry sparked your interest in using click chemistry for your imaging experiments? Learn more about in this webinar: “Live-cell imaging with nanometer probes and tags”.
With the advancement of superresolution microscopy techniques such as STED or MINFLUX, the size of the label plays an increasingly important role. The clickable system of bioorthogonal reaction groups and tetrazine-containing fluorophores – such as our abberior LIVE click probes – expands the toolbox of small-format labeling methods for live-cell imaging.
In this webinar, Dr. Ivana Nikić-Spiegel, group leader at the University of Tübingen, gives us a deeper insight into these nanometer-sized tags.
Dr. Gražvydas Lukinavičius, group leader at the Max Planck Institute for Biophysical Chemistry in Göttingen will show the influences of small changes at the ligands and organic fluorophores, leading to increased binding affinity and biocompatibility.
Watch the recording of the webinar from September 15, 2021
Our course at the European Molecular Biology Laboratory (EMBL) taught users everything they need to know to use our STED and MINFLUX systems to the max. Distinguished external speakers together with abberior’s own application team gave a thorough understanding of the theory, and in practical courses and Q&As, we honed the skills required to take advantage of the most recent advances.
Topics included sample preparation, 3D imaging, live-cell, tissue and fluorescence lifetime STED imaging, MINFLUX tracking, and data analysis for both technologies.
A big thank you to every participant, speaker, organizer, and supporter!
We demonstrate two-color STED imaging of living cells using various pairs of dyes. To this end, we combine click-chemistry-based protein labeling with other orthogonal and highly specific labeling methods, enabling long-term STED imaging of different target structures in living specimens.
Publication
Carola Gregor, et al.: “Two-color live-cell STED nanoscopy by click labeling with cell-permeable fluorophores”, bioRxiv (2022)
Details >
Germany is known for its innovative and high-performing SMEs as the basis of overall economic success. In 2021, there were around 6.1 million small- and medium-sized enterprises in Germany, which generated a revenue of approximately 5.6 trillion euros.
With these impressive figures, one can be justifiably proud when the renowned business magazine WirtschaftsWoche places you among the 100 most innovative SMEs in Germany.
Our thanks go to all our employees – we couldn’t have done it without you.
WirtschaftsWoche ranks abberior
Take a look at our booth at MAF 2022 in Gothenburg and we’ll take a look at your sample.
Just label your sample beforehand, come by and see it immediately super crisp and in super resolution. Thanks to our easy-to-use and rock-solid STEDYCON, you can use STED microscopy anytime, anywhere.
Sign up for a test session and we’ll send you our STED dyes in advance. Don’t miss the chance to measure your samples live and on site.
By the way: We will raffle a dye set among all participants.
Congratulations to our co-founder Stefan Hell on his acceptance into the Order Pour le mérite.
“Admission to the Order is one of the highest honors that can be bestowed on a scientist or artist in Germany. The association of artists and scholars was founded in 1842 by Prussian King Frederick William IV and revived in 1952 by German Bundespräsident Theodor Heuss. It is under the protectorate of the Bundespräsident. The first chancellor of the order was the naturalist Alexander von Humboldt.” BPA, 22.07.2022
Press releases:
Presse- und Informationsamt der Bundesregierung (BPA)
Details >
Max-Planck-Institut, Göttingen
Details >
Have a look at these two independent publications on MINFLUX tracking of motor proteins kinesines moving along microtubules.
Congratulations to @JonasRies Lab, @Stefan_W_Hell and @StefanHellLabs.
Publications
Jan O. Wolff, et al.: “MINFLUX dissects the unimpeded walking of kinesin-1”, bioRxiv (2022)
Details >
Takahiro Deguchi, et al.: “Direct observation of motor protein stepping in living cells using MINFLUX”, bioRxiv (2022)
Details >
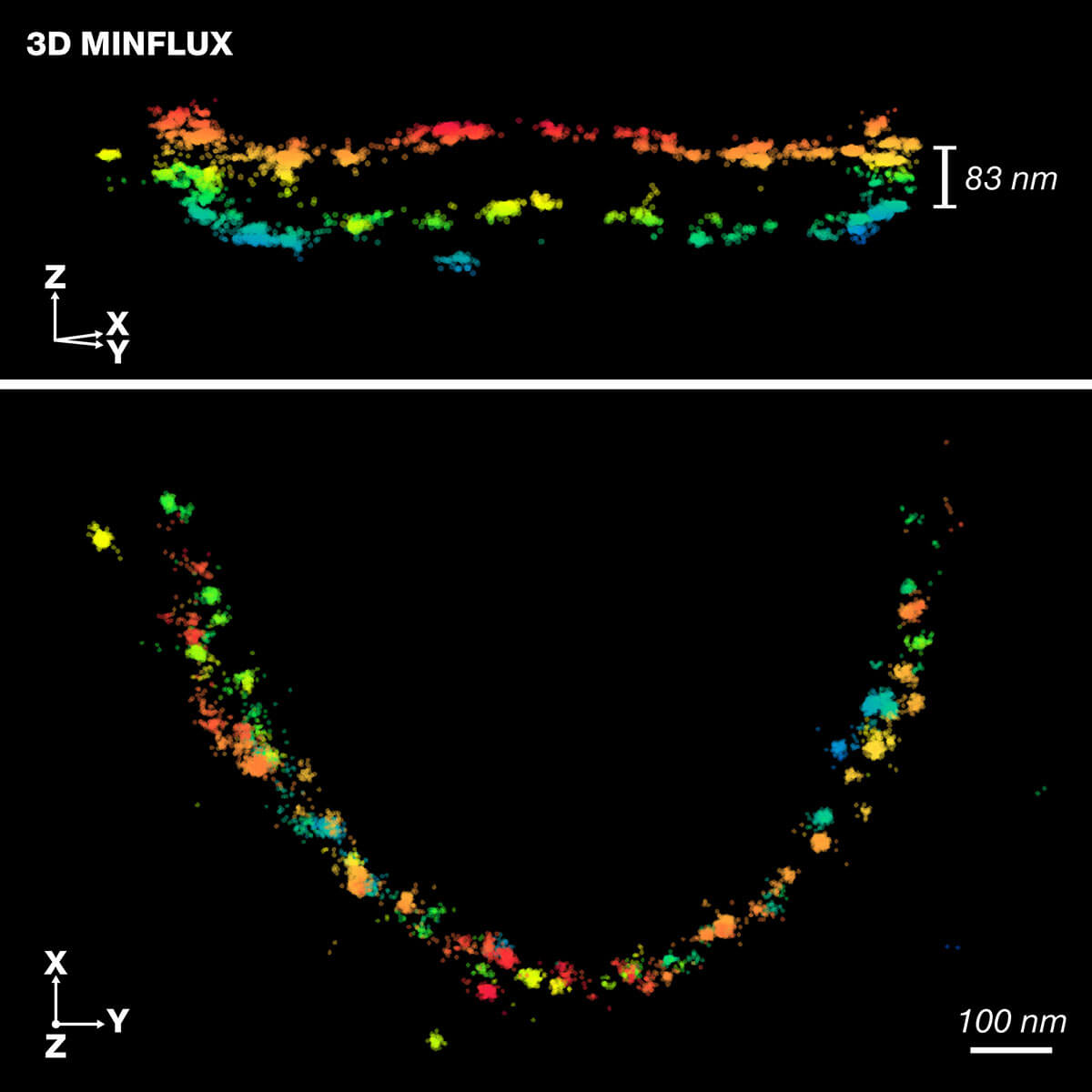
Scientists at the Institute for Auditory Neuroscience, UMG, the Max Planck Institute for Multidisciplinary Sciences and abberior instruments apply high-resolution 3D-MINFLUX technology for precise 3D representation of the molecular organization in the active zone of rod photoreceptor cells. Published in Science Advances.
Running is a lot of fun for everyone: This becomes clear at the 32. Göttinger Altstadtlauf. After a two-year break from the Corona, several thousand athletes start the race. The abberior running team participated in the Company Cup this year.
We were not aiming to be the first to cross the finish line 🙂 It was about being strong together as a team, achieving something together, having fun, and toasting our achievement with a smile on our faces at the end.
This is a wonderful team spirit!
Live-cell fluorescence microscopy is an indispensable tool in biomedical and biophysical research, allowing the study of dynamic processes in living systems. We are excited to present our new copper-free clickable dyes, a minimal-sized tag for biorthogonal live-cell labeling.
Our new abberior LIVE click tetrazine-based probes expand the toolbox of super-efficient organic dyes for multicolor live-cell imaging. Specific labeling of intracellular targets using abberior LIVE probes for DNA, actin, tubulin, SNAP, click, and STAR membrane, making long-term live-cell imaging at extremely low nanomolar probe concentrations possible.
This unique combination of tags takes live-cell imaging to the next level and will make super-resolution imaging of living systems a routine, easy-to-use visualization method and paves the way for a deeper understanding of the nanoarchitecture of life.
Do you often wonder what strategies are available to optimize your multicolor STED experiment?
Listen to our speakers Elisa D’Este, head of the optical microscopy facility at the MIP for medical research, and Florian Grimm Business Developer at abberior. See how different labeling techniques like indirect immunofluorescence, nanobodies, direct labels, membrane probes, or self-labeling enzymes can be combined to achieve colorful images at the highest resolution in living or fixed samples.
Watch the recording of our last webinar on July 7, 2022, 15:00 (CEST)
In November last year we celebrated the foundation laying ceremony for abberior’s new headquarters. The new building is being constructed at Göttingen’s North Campus in the direct vicinity of our founding location and previous HQ under the roof of IFNANO. In addition, the Max Planck Institute for Multidisciplinary Sciences (MPI-NAT), a close partner in research, is not far away as well. The proud construction workers were joined by abberior’s founding team, the organizers behind the new building, and the new head mayor of Göttingen, Petra Broistedt, to celebrate the occasion. In her speech on the occasion, she emphasized how important abberior and its contribution to the further development of the world of microscopy is both for researchers worldwide and for Göttingen as a location.
Now it won’t be long until the grand opening and the relocation. We are already excited!
Until then, we will keep you up to date with the progress on our social media pages (Twitter, Instagram, LinkedIn, Facebook). And while you’re at it: Why not have a look at our current job postings and save yourself a seat in our new headquarters?
“Invented, developed and built in Göttingen: Thanks to the discovery of Nobel laureate Stefan Hell, scientists around the world are seeing things through abberior microscopes that they have never seen before – and can achieve decisive breakthroughs in research.”
Explore abberior’s new lifetime toolbox TIMEBOW with Julia & Gero. See how lifetime and STED superresolution imaging come together easily with our phasor-based tools. MATRIX detection adds unique lifetime imaging modes and extra clarity to the mix.
Watch the recording of our webinar on April 21, 2022, 15:00 (CEST)
We are very pleased to announce that our founder Prof. Stefan Hell will meet you online next Thursday at iCANX SUMMIT 2022. He will introduce the revolutionary localization principle MINFLUX and present his latest imaging technology MINSTED.
You can click here > or scan the QR code to watch the talk online.
Title of talk:MINFLUX and MINSTED enable molecule-level resolution in fluorescence microscopy.
Date and time: Thursday, April 28, 2022, 2:00 PM (CEST).
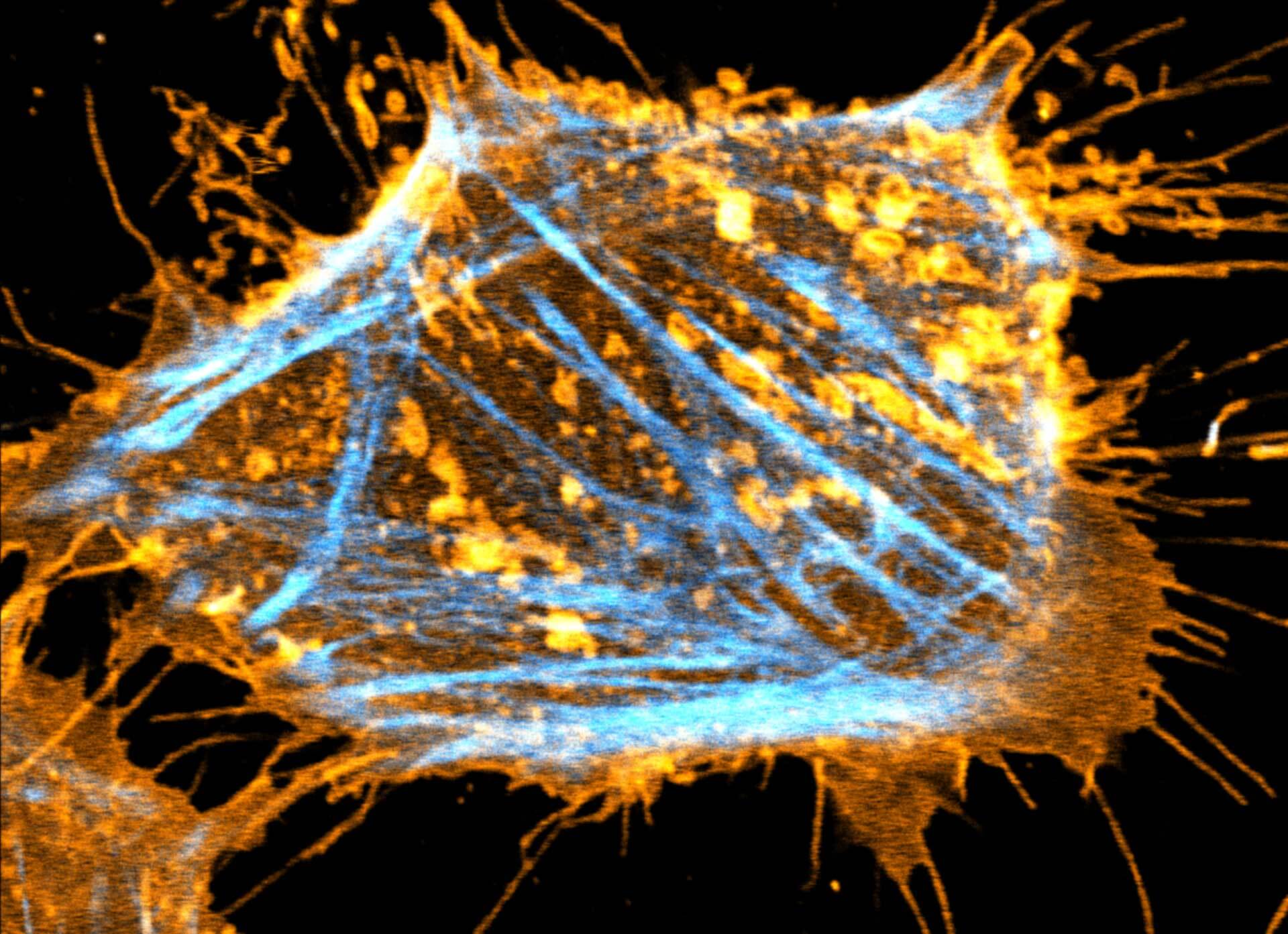
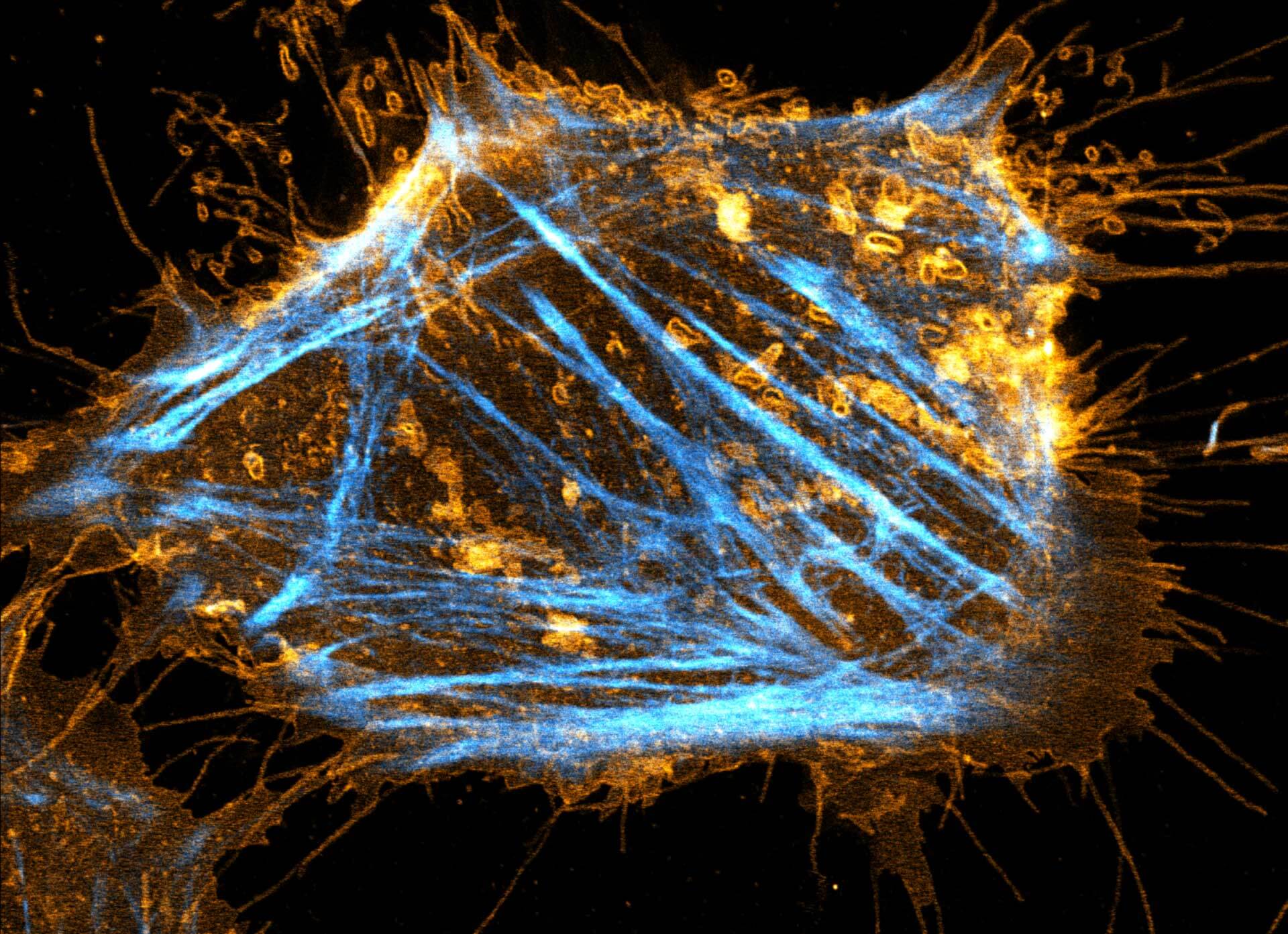
Description
Two color live-cell STED and confocal image of a mammalian cultured cell stained with abberior STAR RED membrane (orange) and abberior LIVE 590 actin (cyan).
Live-cell fluorescence microscopy is an indispensable tool in biomedical and biophysical research, allowing the study of dynamic processes in living systems. We are excited to present our new membrane dyes, that visualize the plasma membrane of living cell!
abberior STAR membrane probes profit from a special linker designed to minimize the impact on the membrane to maintain the overall mobility and diffusivity. Try the new abberior method of cell membrane labeling. In combination with our abberior LIVE dyes, you get a toolbox of super-efficient probes, that provide specific labeling of intracellular targets and outer cell membrane, making long-term live-cell imaging at extremely low nanomolar probe concentrations possible.
This unique combination takes live-cell imaging to the next level and will make super-resolution imaging of living systems a routine, easy-to-use visualization method, paving the way for fundamental biological discoveries.
The EMBL Imaging Centre and abberior Instruments GmbH extend their collaboration. After a successful first phase of three years, both parties agreed to continue the joint development of technology and applications for the next generation MINFLUX.
MINFLUX is the latest breakthrough in light microscopy, capable of observing biological structures with unprecedented temporal and spatial resolution. A particularly exciting area for groundbreaking biological applications is the structural biology of living systems. Here, the envisioned unique benefit of MINFLUX will be to observe the critical conformational changes in real-time at the nanoscale that allow molecular machines to perform their function.
EMBL Imaging Centre:
With 27 member states, the European Molecular Biology Laboratory (EMBL) currently employs more than 1800 people with more than 110 independent research groups and service teams covering the spectrum of molecular biology at six sites in Barcelona, Grenoble, Hamburg, Heidelberg, EMBL-EBI Hinxton, and Rome. The EMBL Imaging Centre is the latest addition to the EMBL headquarters in Heidelberg. It offers the most advanced high-resolution electron and light microscopy technologies, including new and in-house developed methods that are not yet commercially available. The EMBL Imaging Centre builds on EMBL’s strength in imaging technology development and nearly five decades of experience in world-class service provision for life science research. This will encompass external access to novel, cutting-edge superresolution technology, which includes MINFLUX technology, in both commercial and pre-commercial configurations and performance. Each year up to 150 visiting scientists will be able to visit the new center to access the very latest imaging technologies. EMBL’s highly qualified experts will provide the visitors with tailored support from sample preparation to data analysis. An additional aim of the center is to develop the next generation of imaging technologies for life science applications in close cooperation with industrial partners.
abberior Instruments:
abberior Instruments and it’s sister company abberior were founded in 2012 as spin-offs of the Max-Planck Institute for Biophysical Chemistry in Göttingen and the German Cancer Research Center in Heidelberg. The mission of the founders around Nobel laureate Prof. Dr. Stefan Hell is to make groundbreaking developments in high-resolution light microscopy available to biological and medical researchers. The focus is on both rapid implementation and, at the same time, simple and broad applicability. Numerous awards, including the TOP100 Innovation Award 2021, the Innovation Award of German Science, and the Focus Growth Champion 2019, highlight the success of the abberior group. Meanwhile, abberior Instruments has offices in Europe, America, and Asia and has put up hundreds of microscopes in more than 30 countries with 100 very talented people working all around the globe.
This short video covers fluorescence lifetime analysis and separation using phasor plots with confocal and STED superresolution. Additionally, Julia and Gero demonstrate background subtraction with MATRIX and they show how to achieve high resolutions at only 2% STED laser power using lifetime data.
Lifetime imaging was never so easy and TIMEBOW and MATRIX array detection are a winning combination – only available from abberior. See for yourself!
abberior, the leading provider of STED and MINFLUX super resolution microscopes, announces a new strategic partnership with OptoFeM. This cooperation empowers both parties to deliver the highest levels of quality, service, and application support covering whole Abberior Instruments product portfolio to customers in China.
The collaboration already started in 2019 and is now lifted to the level of strategic partnership, being developed into a highly intensive, large-scale collaboration in the fields of application support, sales, and service.
“We are very happy to work so efficiently, seamlessly and successfully together with our partner OptoFeM. We are jointly accelerating the adoption of our abberior super resolution technology, and we can feel that our brand is getting high traction in China. It is very fascinating to see how quickly and professional scientific research is developing there. Our quality and the abberior super resolution performance is highly appreciated by our customers in China.” says Dr. Gerald Donnert, CEO of abberior Instruments GmbH.
“OptoFeM and abberior have joined hands in strength and in-depth cooperation. Relying on abberior’s outstanding optical imaging products and OptoFeM’s sound sales and service system together we are developing the Chinese optical imaging market. We look forward to providing more outstanding products and service to Chinese customers.” says Mr. Qi Dong, CEO of OptofeM.
Congratulations to our founder Stefan Hell, who shares the Werner-von-Siemens Ring 2022 with the main scientific forces of BioNTech, Uğur Şahin, Özlem Türeci, Christoph Huber and Katalin Karikó.
Stefan is honored for being the first to show that the diffraction barrier of light microscopy can be broken. His STED microscopes achieve resolutions up to 20 nm, about a hundred times sharper than conventional instruments. Other physicists soon realized the potential of the principle and developed similar techniques. Together, these quickly arrived in life science research and revolutionized light microscopy by opening the door to the visualization of cellular structures in unprecedented detail.
Among many prizes, Stefan Hell received the Nobel Price in Chemistry 2014. Since then, he and his fellow colleagues have not rested and pushed the achievable resolution to the molecular scale with MINFLUX. This microscope can discern molecules even if they are just one nanometer apart, and they can track their position hundred times faster than before.
Facts aside, of course, we at abberior know all of this, since we have the privilege of looking through these spectacular instruments every day when we build, develop, and market them. And so do our 100 employees in Europe, China, North America, and thousands of customers using our instruments all over the world.
In abberiors’ constant quest to provide researchers with revolutionary tools for high-quality confocal and super-resolution microscopy, we present our newest development: TIMEBOW lifetime imaging.
TIMEBOW provides a straightforward way to take advantage of the lifetime information that is carried by every photon from the sample. Instead of looking at intensity only, discarding this information, TIMEBOW exploits it and advances your imaging with and without super resolution. Within seconds, lifetime variabilities are turned into color-coded images, dyes become unmixed and resolution is increased.
In combination with our groundbreaking MATRIX array detector, TIMEBOW is especially powerful – TIMEBOW boosts the resolution and MATRIX removes the background. What more could you ask for?
2021 has been a tumultuous year for all of us, full of small and big adjustments. Despite the challenges, abberior can also look back at many exciting developments. With the year slowly coming to its and, we would like to take a moment to focus on what made 2021 matter.
We keep on breaking barriers: In our quest to continuously provide the world’s first-class resolution microscopy, we introduced MINFLUX 3D this year. This technology allows our MINFLUX platform to achieve single-digit nanometer resolution in all three spatial dimensions – a level that is unachievable by any other product on the market. Our assortment in dyes and labels has also been greatly expanded. Especially in the sector of live-cell microscopy and Long-Stokes shift, we advanced the toolkit for scientists by developing new fluorophores.
Innovation is the name of our game: This year we participated for the first time in the TOP 100 competition for innovation in German small and medium enterprises. After passing their test, a data driven performance analysis, we received the prestigious TOP 100 seal and even took second place in our size class. The German Physical Society honored us with their Award for Technology Transfer. The award highlights the work that goes into making an invention available.
Expanding in times of a pandemic: The coronavirus continued to dictate many aspects of our daily lives. Lots of energy and time were invested to create the safest possible work environment for our employees. While all this was happening, abberior surely did not stop growing. For example, this year saw the establishment of our second office in the USA and our first microscope being delivered to Russia. Special attention was given to the Chinese market: we established our first local office with service and sales employees in Beijing and launched a custom Chinese website. Our central office in Göttingen kept growing as well, with our current facilities running out of space. Luckily, relief is on the horizon – literally – with our new headquarters starting development in a brand-new building right around the corner.
These days, team spirit and solidarity are especially important, and we are happy to have achieved so many great things together. And we will continue to do so in 2022, with many exciting things to come. But for now, we wish you and your loved ones a restful holiday season and a safe start into the New Year.
The German Physical Society (DPG) has awarded the Technology Transfer Prize to abberior, the Max Planck Institute for Biophysical Chemistry and Max Planck Innovation GmbH. The three institutions receive the prize for the successful industrial implementation of research results in the field of high-resolution light microscopy. The award recognizes the many steps that go into the realization of innovative discoveries into an entrepreneurial setting.
abberior emerged in the early 2010s from Stefan Hell’s NanoBiophotonics department at the MPI for Biophysical Chemistry. Max Planck Innovation GmbH, as a link between science and industry, accompanied the founding of the company and licensenewd out the findings in the field of fluorescence microscopy to the then start-up.
Gerald Donnert, our CEO, is delighted with the recognition and at the same time sets clear goals: “In the end, our microscopes are tools that can be used to make fundamental discoveries in biology and medicine that will benefit everyone in the future. At abberior Instruments, we want to provide researchers from around the world with the sharpest and most powerful fluorescence microscopes that can be built today. This advances industrial and academic research and helps to identify the systematics of disease or life on a molecular scale.”
Our main goal at abberior is to push beyond barriers with new discoveries and to make those discoveries accessible to scientists around the world. That in itself carries lots of innovative spirit. Add to that a skilled young team, a constant stream of new inventions and the ambitious energy of a start-up and you get one of the most innovative enterprises in Germany.
Now the winners of the prestigious TOP 100 award were announced in a virtual conference by mentor Ranga Yogeshwar. The innovation competition, which is being held for the 28th time, analyzed all applicants on the basis of more than 100 performance indicators in five categories before awarding the seal. Among companies of similar size abberior Instruments took the second place, with our particularly impressive performance in the category “Innovation Success” being highlighted.
The scientific management is organized by Prof. Dr. Nikolaus Franke. As founder and director of the Institute for Entrepreneurship and Innovation at the Vienna University of Economics and Business, Franke is one of the leading innovation researchers internationally. Project partners are the Fraunhofer Society for the Promotion of Applied Research and the BVMW association of small and medium-sized enterprises.
It’s a new era for abberior. Last Friday, we celebrated the laying of the foundation stone for our new headquarters in Göttingen. The new building will be located in the direct vicinity of the researchers at the North Campus of the University of Göttingen and the Max-Planck-Institute for Biophysical Chemistry. As co-founder and Nobel prize winner Stefan W. Hell explained in his speech: “What better way for researchers at Göttingen University to know that the world’s sharpest microscopes are literally built a few hundred meters around the corner. Or that one or two phone calls are enough to be able to bring along a sample and make the crucial measurement for a research paper. You won’t find that anywhere else.”
The project is the result of a collaboration between our companies, the GWG, an investor for urban business development, the city of Göttingen and the University of Göttingen, who supplied the building area. In total, 14 million euros will be invested in the seven-story building to create research, production and office spaces for 200 employees. The building is planned to be finished in the second half of 2022. Not a day to soon, adds co-founder and CEO Dr. Gerald Donnert: “Currently, we hire 30 new people each year.”
Until then, we will keep you up to date with the progress on our social media pages (Twitter, Instagram, LinkedIn, Facebook). And while you’re at it: Why not have a look at our current job postings and save yourself a seat in our new headquarters?
China is very important for us and for the world! We are proud to announce that abberior is actively expanding the Chinese market. Users around the world can now directly benefit from the “latest STED and MINFLUX systems with the highest spatial resolution in fluorescence microscopy”, as Prof. Stefan W. Hell stated.
abberior has opened the first office in Beijing and is looking forward to welcoming guests. Here, our experienced scientists and engineers will give customers vivid system presentations and demonstrations on site or remotely.
To make access to our technology as easy as possible, we launched our Chinese website today. With just a few clicks, scientists and researchers can find key product information, learn how they can benefit from patented abberior technologies, have all company news at their fingertips, and get direct access to local technical support.
We are convinced that direct and barrier-free communication with our customers, strengthens mutual trust and cooperation and will be a guarantee for future success on both sides.
abberior and the European Molecular Biology Laboratory (EMBL) have launched a collaboration on the application of MINFLUX in biology. The goal of this agreement is to combine forces regarding further development of the method, and to apply superresolution MINFLUX microscopy to living systems.
To this end, abberior installed a MINFLUX system at the EMBL Imaging Centre in Heidelberg, Germany in August 2020. This nanoscope will be used for developing imaging workflows and for targeting a broad range of biological questions. After successful proof-of-concept applications, the system will also be available for external users in the framework of the new EMBL Imaging Centre.
“This collaboration will allow us to apply MINFLUX to highly relevant questions in biology and to push the limits of this technology. In the near future, we will to adapt MINFLUX microscopy to operate in living systems.” said Christian Wurm, Head of the abberior application team.
The abberior MINFLUX system is the newest addition to the abberior product portfolio. It is the first commercialized microscopy system that provides true molecule-size resolution and holds the world-record in temporal resolution for single particle tracking.
abberior has opened its new office in Bethesda, Maryland, USA. The new site houses conference and showrooms, demo rooms and offices, as well as a supply for replacement parts.
“abberior Instruments America is excited to have opened our new office in Bethesda Maryland located less than 1 mile from the National Institutes of Health main campus and within a short drive of key research centers at Johns Hopkins and Howard Hughes’ Janelia Farms.” says David Melton, CEO of abberior Instruments America.
Our US staff and two microscopes, a FACILITY line and a STEDYCON, are looking forward to our customers, many interesting discussions, live and remote demos, and exceptional images.
Using an abberior STED-FCS, a team from four European countries, comprising Christian Eggeling, Leibniz IPHT & Friedrich Schiller University Jena and Delphine Muriaux, Montpellier Infectious Disease Research Institute, discovered the existence and composition of a lipid-based platform for HIV virus assembly. These results provide novel molecular insights on HIV virus assembly and are a starting point for the development of new antiviral drugs.
Read here: “HIV-1 Gag specifically restricts PI(4,5)P2 and cholesterol mobility in living cells creating a nanodomain platform for virus assembly”, Science Advances
In this article, Prof. Stefan Hell talks about his professional career, how he came to develop STED microscopy in the early nineties, and how our company was started and where it’s heading.
It’s a fantastic read about the early times of superresolution all the way up to now, with many interesting details about why things unfolded in a particular way. Don’t miss it if you’re interested in the history of STED, abberior, or Stefan Hell!
Further Information:
Website Max Planck
Full Article (German)
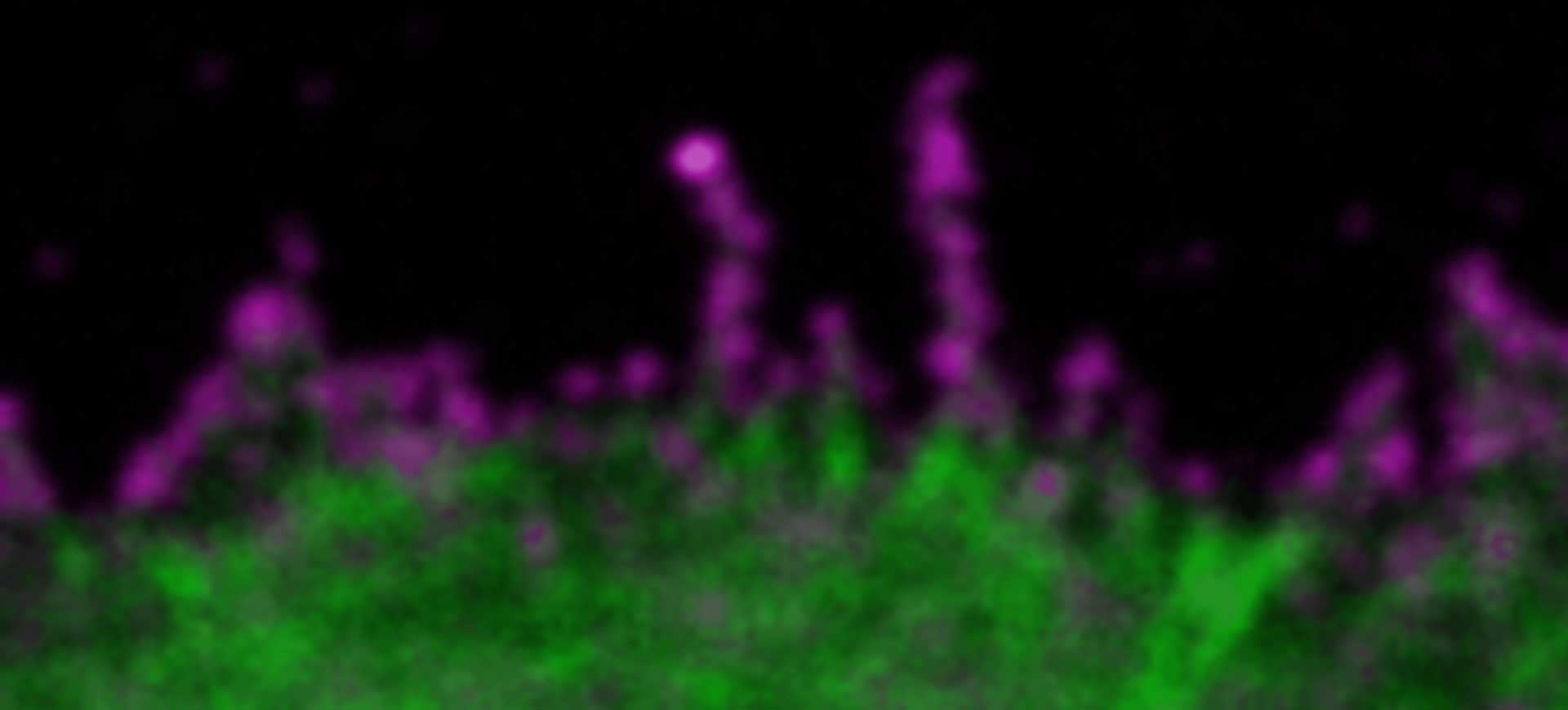
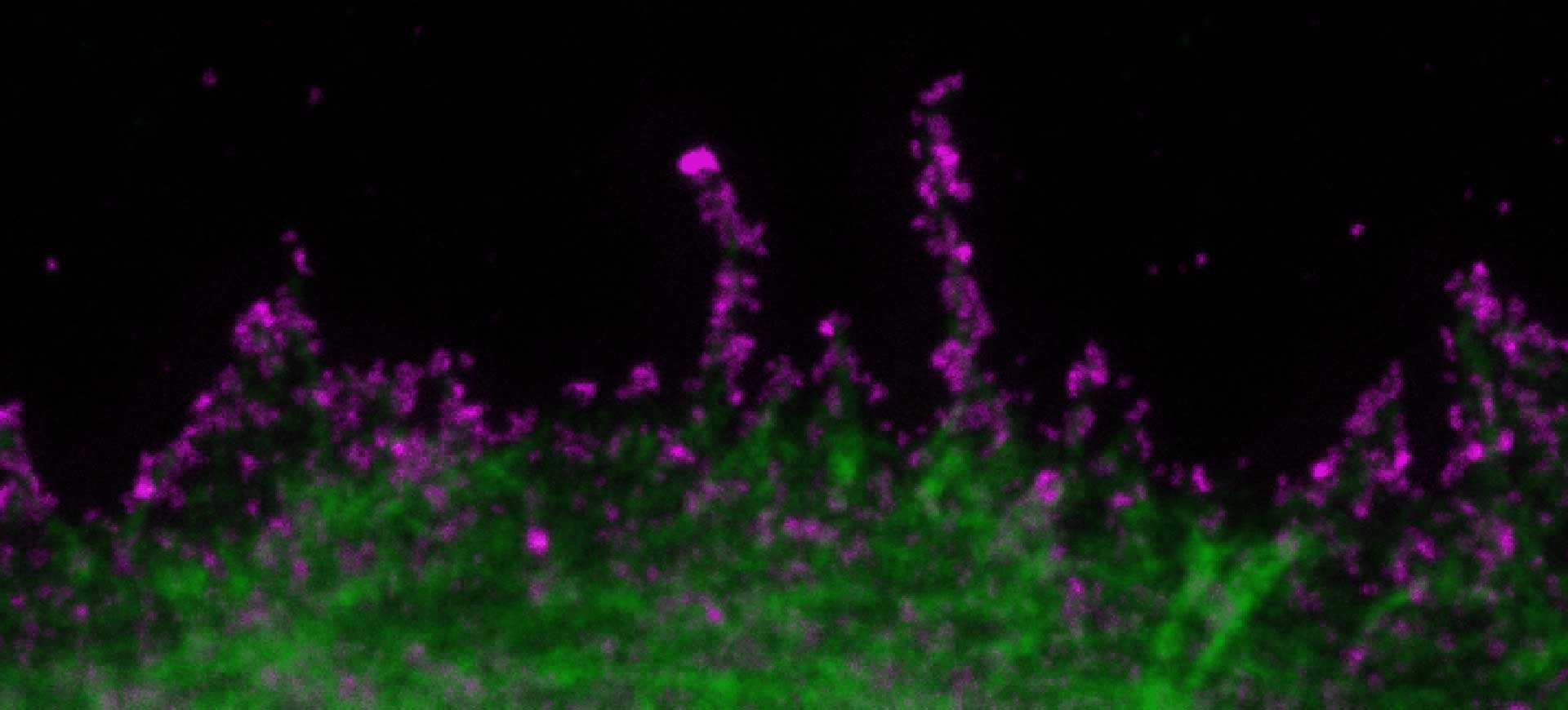
Description
SARS-CoV2 (“Corona”, magenta, abberior STAR RED) and cellular actin (green, abberior STAR ORANGE Phalloidin) in VeroE6 infected cells.
Copyright: MDVA team/CNRS Montpellier, France (D. Muriaux, C. Favard, M.-P. Blanchard @MRI)
Scientists in France have successfully used an abberior STED system and abberior dyes to shoot images of SARS-CoV2 (“Corona”, magenta) infecting VeroE6 cells.
Understanding the precise structure of this virus and the processes during and after infection are crucial to finding a cure or vaccine, ending lockdowns all around the world. Standard light microscopes are not of much help, since they are limited by diffraction (approx. 200 nm), while the virus particles have a much smaller diameter at about 100 nm. Because of this, SARS-CoV2 and important cellular structures that it attacks only show up blurry in standard, e.g. so-called confocal microscopes.
In the past, electron microscopy would have been used, but sample preparation for this instruments in tedious and they cannot image living matter, which is nevertheless vital to unraveling the workings of a corona infection on the cellular level. This is where STED microscopy comes into play: with a resolution 5-10 times below the diffraction limit, and much weaker constraints regarding sample preparation, it’s going to be an important tool for finding drugs against corona and similar viruses.
Copyright: MDVA team/CNRS Montpellier, France (D. Muriaux, C. Favard, M.-P. Blanchard @MRI)
abberior, the leading STED solution provider from the inventors of STED and Nikon Instruments Inc., innovator of advanced microscope systems today announced a strategic partnership to accelerate adoption of easy-to-use STED technology in the US. As part of this collaboration, abberior’s compact STEDYCON system will be distributed by Nikon.
Inquiries for abberior Instruments America: David Melton | CEO | 561.972.9300 | info@abberior-instruments-america.com
Media Inquiries for Nikon Instruments Inc.: Kristina Corso | Communications Specialist | 212.931.6189 | kcorso@hotpaperlantern.com
Further Information
When we started our business in 2012 in a small office ‘garage’ in Göttingen, our vision was to become the technically leading manufacturer for superresolution microscopes worldwide. With currently around 50 employees in Heidelberg (Germany), Jupiter (Florida, USA) as well as in Basel (Switzerland), we have turned into a globally recognized maker of superresolution light microscopes, selling in more than 35 countries. Due to the excellence of our highly selected and skilled team we have been able to grow with remarkable speed, and now we’re officially recognized as a growth champion!
With fourth place in the category “manufacturing industry”*, and ranked number 133th of 500 companies nationwide, we are very proud to be part of the “Wachstumschampions 2019”. Our average growth between 2014 and 2017 was almost 60% per year.
The Focus ranking lists companies that have achieved the highest sales growth per year in the last three years. Statista sieved through thousands of companies with above-average growth from more than 2 million commercial register entries, and invited many more to apply for the 2019 Growth Championships. The companies now listed in the Focus magazine’s top list are the ones that received the “Growth Champion 2019” award.
Further Information
*without automobile, machinery and plant construction industry