Sample gallery
Fluorescence imaging, whether at confocal, STED or MINFLUX resolution, guarantees unique insights into the function and structure of life at the molecular level. Besides the scientific information content, some sample portraits provide simply beautiful images. Enjoy browsing our sample gallery.
the fine art of science
Description
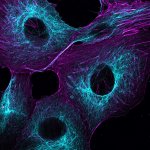
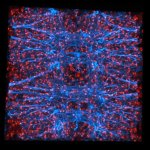
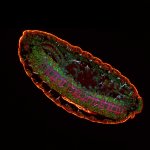
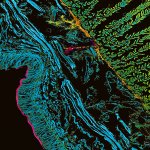
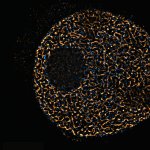
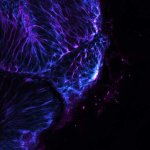
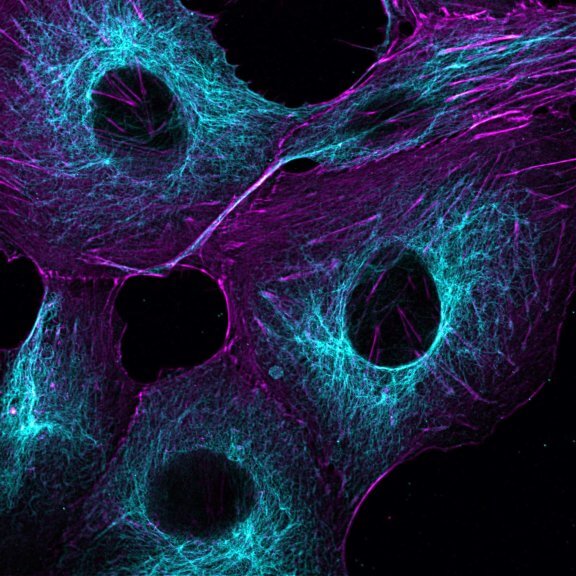
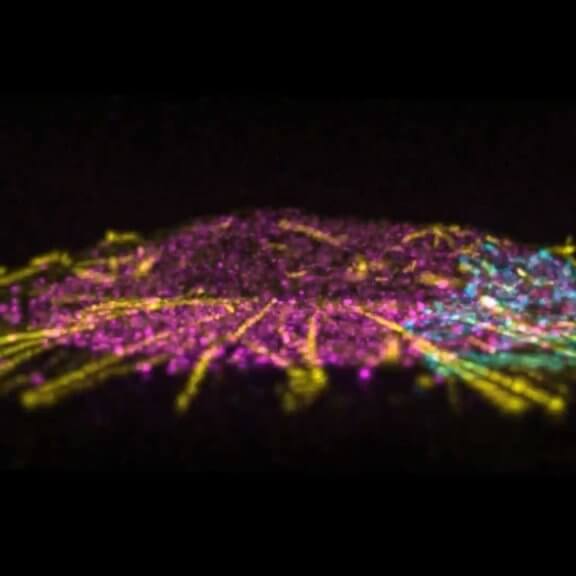
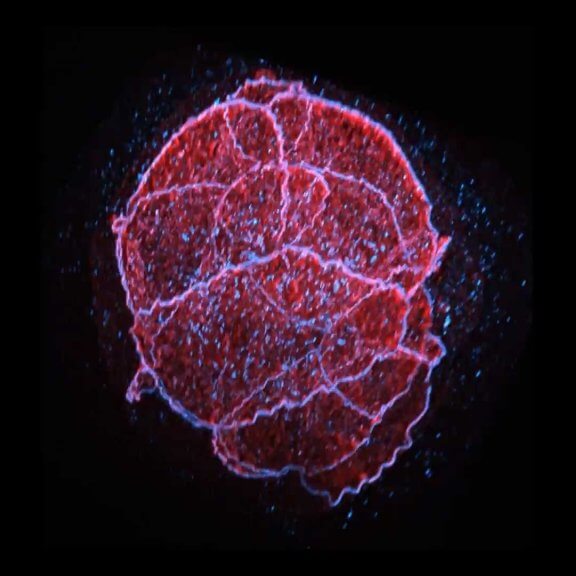
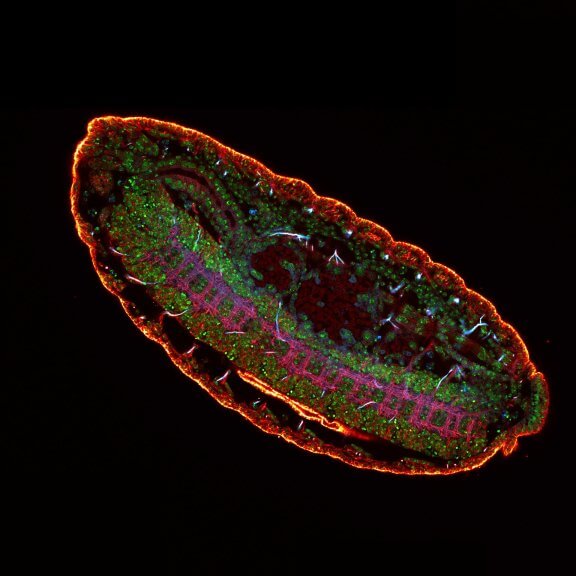
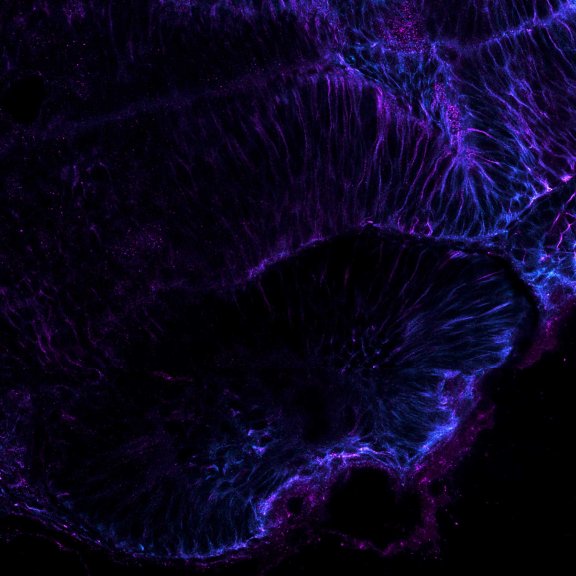
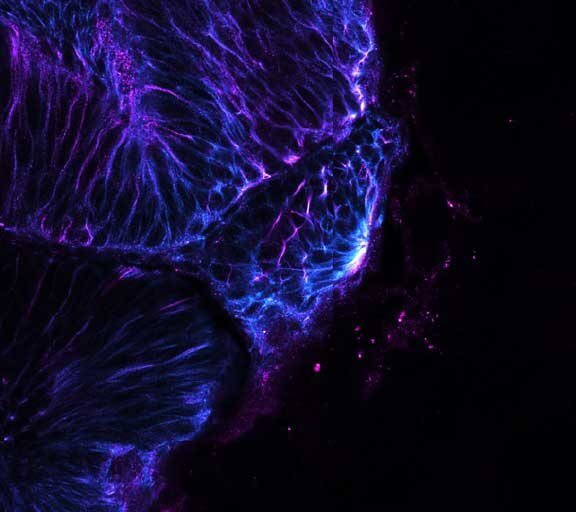
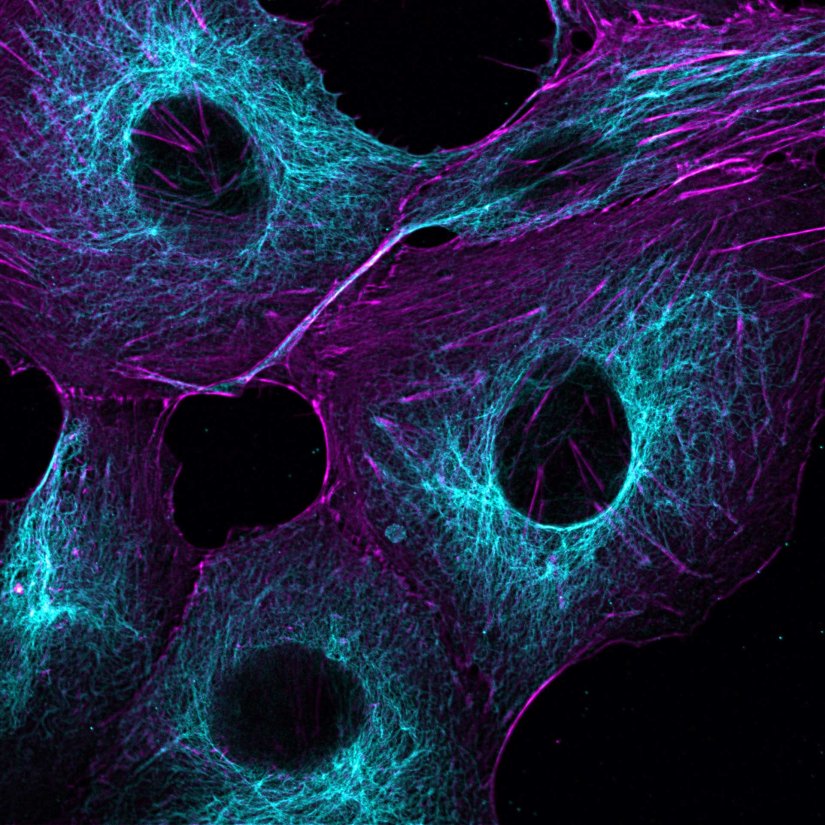
Mammalian cells stained for actin (purple, abberior STAR RED) and vimentin (cyan, abberior LIVE 460L). Differential detection with the MATRIX array detector was used to improve resolution, optical sectioning, and signal-to-background-ratio.
Description
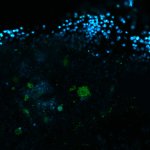
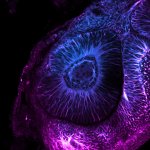
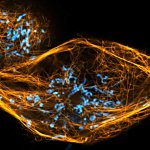
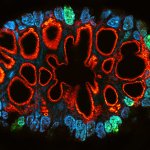
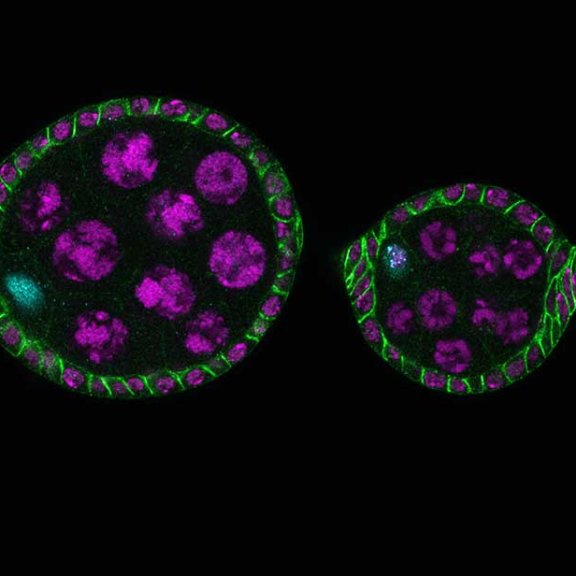
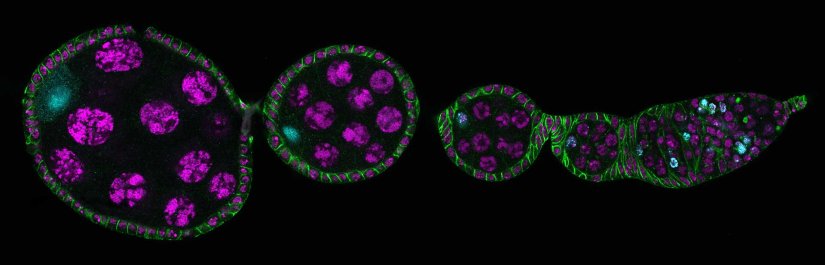
Drosophila ovariole stained for the synaptonemal complex protein C3G (cyan, abberior STAR RED), showing how the complex is formed and disassembled during oocyte development. DNA was stained with DAPI (purple), spectrin with abberior STAR ORANGE (green).
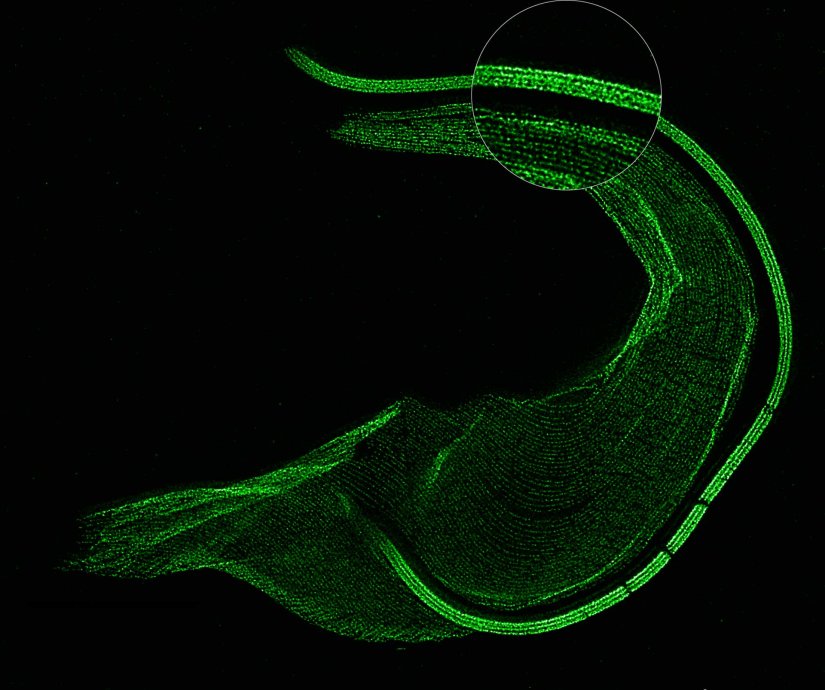
Description
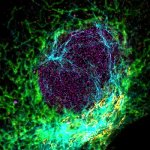
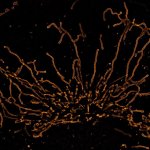
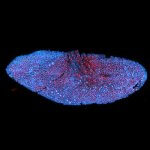
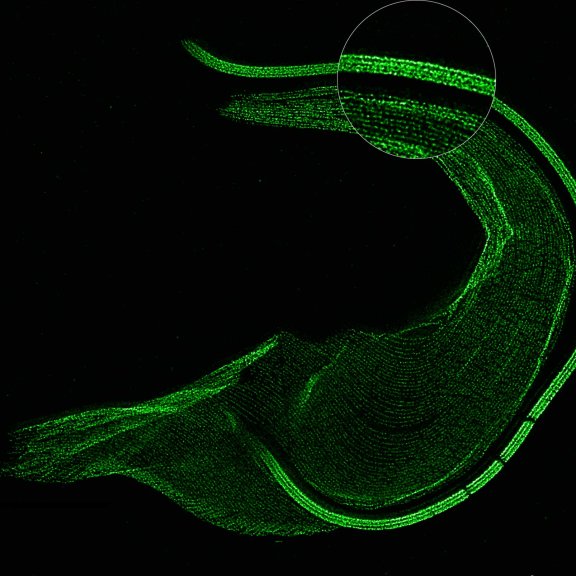
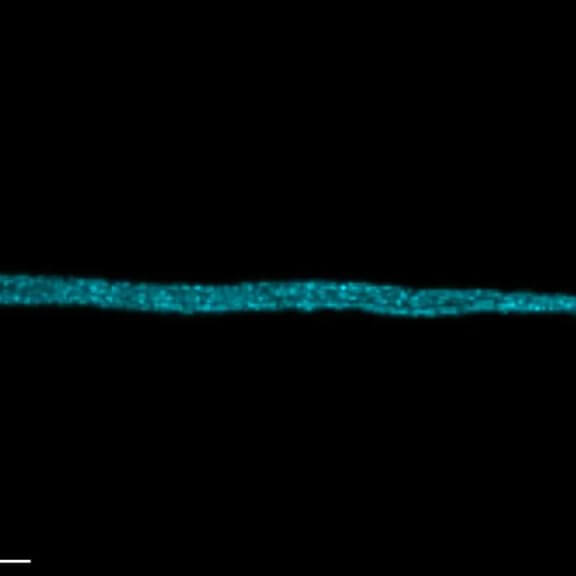
The protozoan Trypanosoma, fixed and three-fold expanded, was stained for tubulin, imaged with STED, and deconvolved with TRUESHARP image boosting. The combination of expansion and STED provides a resolution at which the four tubulin fibers of the flagellum are clearly visible. Scale bar refers to expanded specimen.
Sample courtesy: Mélanie Bonhivers, Laboratoire de Microbiologie Fondamentale et Pathogénicité, CNRS / Université Bordeaux, France
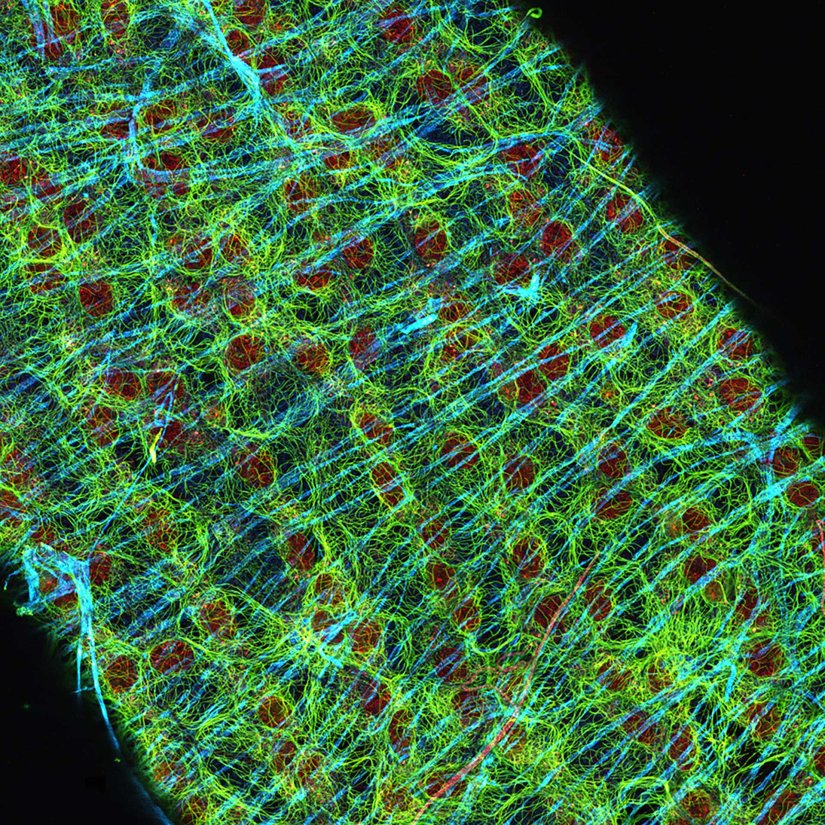
Description
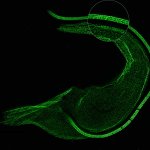
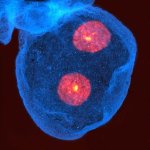
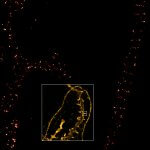
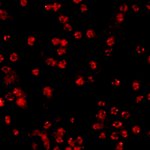
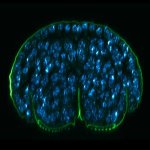
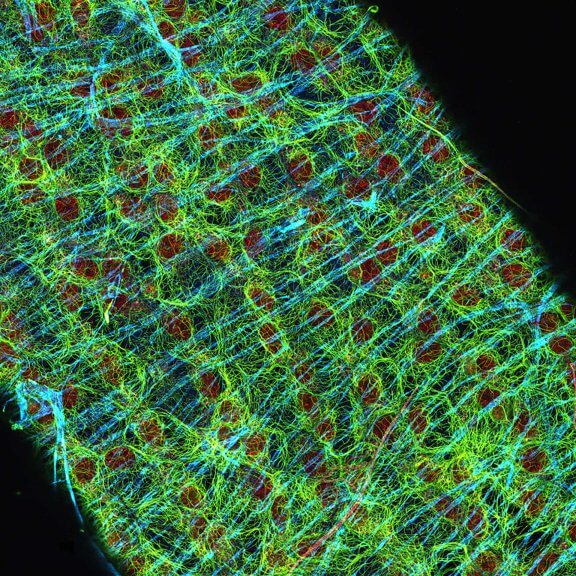
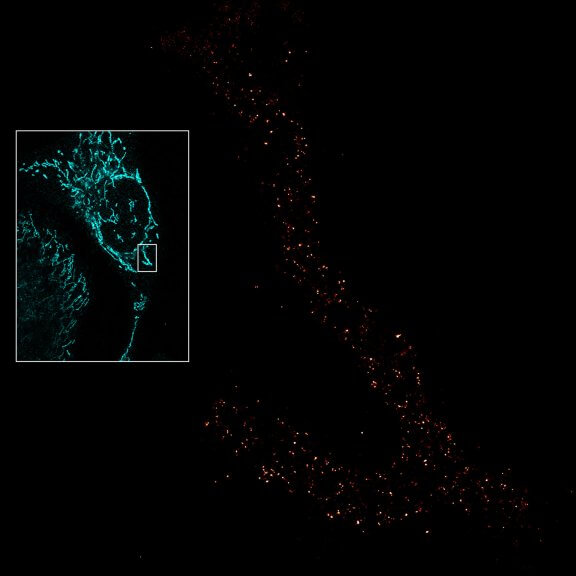
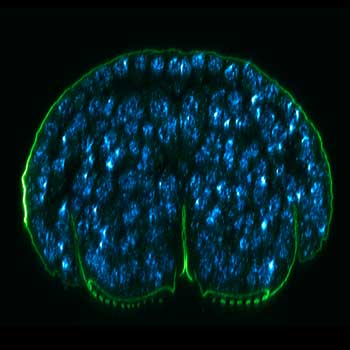
Drosophila egg in the ovary stained for tubulin (green, abberior STAR RED), actin (blue, abberior STAR GREEN), and DNA (red, abberior LIVE 590).
Description
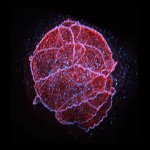
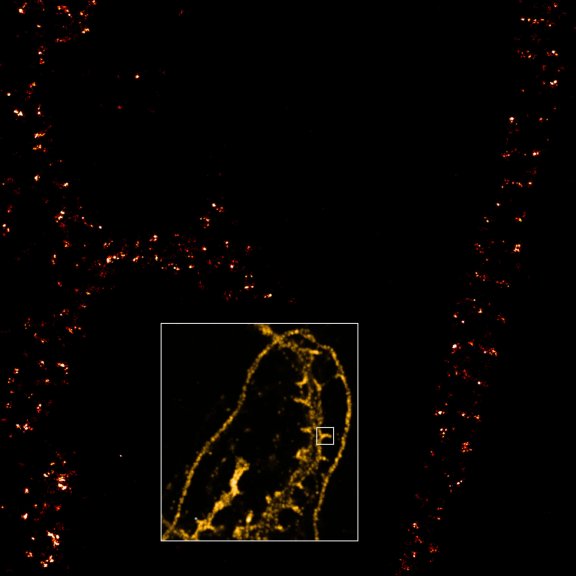
A single 775 nm STED laser was used to visualize this cell’s nuclear pore complex (purple, CF680R), golgi (cyan, abberior STAR RED), and actin (yellow, abberior STAR ORANGE) in superresolution.
Description
A cilium of a fixed RPE-1 cell, gel-expanded and imaged with 3D STED and FLEXPOSURE adaptive illumination. CEP164 was labeled with abberior STAR RED (red) and acetylated tubulin + CCDC41 were labeled with abberior STAR ORANGE (cyan).
Samples courtesy: Dong Kong, PhD & Jadranka Loncarek, PhD. Cancer Innovation Laboratory, NCI/CCR, USA.
Description
Centriolar rosettes in fixed RPE-1 cells, imaged with 3D STED and FLEXPOSURE adaptive illumination. Labeling: CP110 (abberior STAR RED) and SAS-6 (abberior STAR ORANGE).
Sample courtesy: Catherine Sullenberger & Jadranka Loncarek, Cancer Innovation Laboratory, NCI/CCR, USA
Description
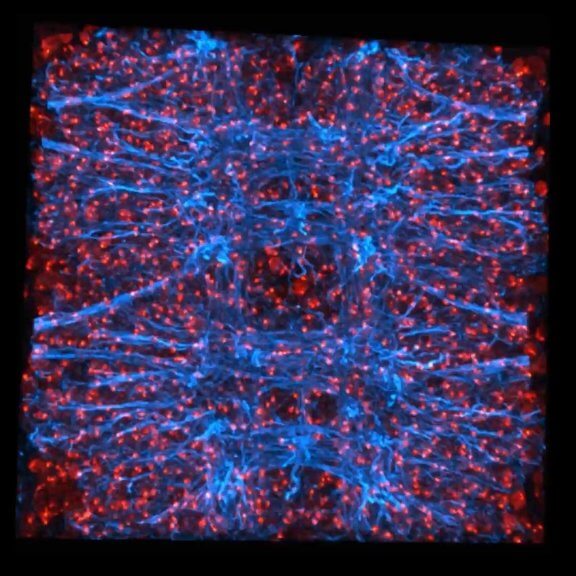
3D STED volume of Drosophila nephrocytes stained for Kirre (cyan, abberior STAR RED), homolog of mammalian Neph1 and a component of the nephrocyte diaphragm, and DAPI (red). Light burden during scan was reduced with FLEXPOSURE adaptive illumination.
Sample courtesy: Konrad Lang, University of Freiburg Medical Center, Germany
Description
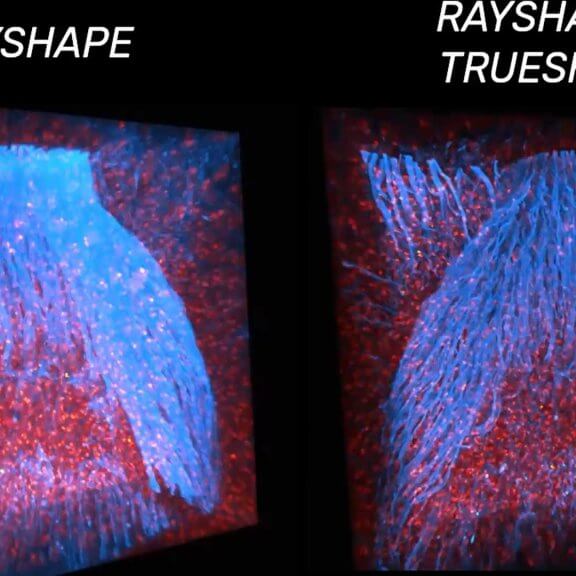
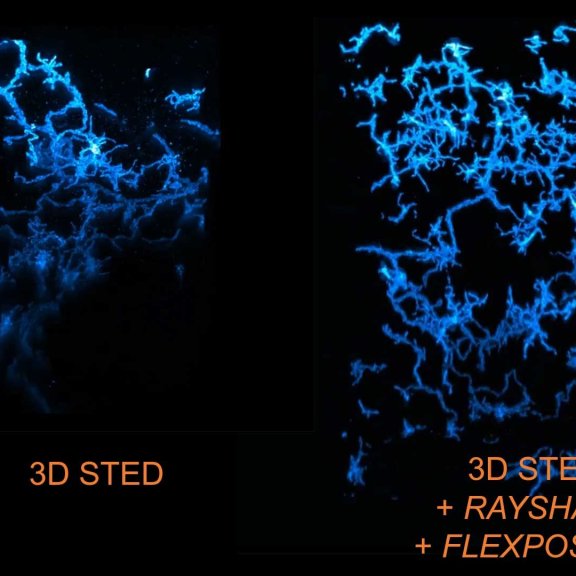
3D STED of microglia and processes labeled for a specific cell surface receptor in neocortical tissue of perfusion-fixed mouse (stained with abberior STAR 635P; unpublished).
FLEXPOSURE adaptive illumination was used to improve signal by reducing light burden and bleaching, RAYSHAPE aberration correction rescued aberration-caused signal loss deep in the sample by adaptive optics.
Sample courtesy: Csaba Cserép and Ádám Dénes, Laboratory of Neuroimmunology, HUN-REN Institute of Experimental Medicine, Budapest, Hungary
Description
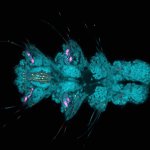
Herpes virus particles 4-5x gel-expanded and imaged with confocal and STED, stained for a viral glycoprotein (orange, abberior STAR RED) and tegument protein (cyan, abberior STAR 580).
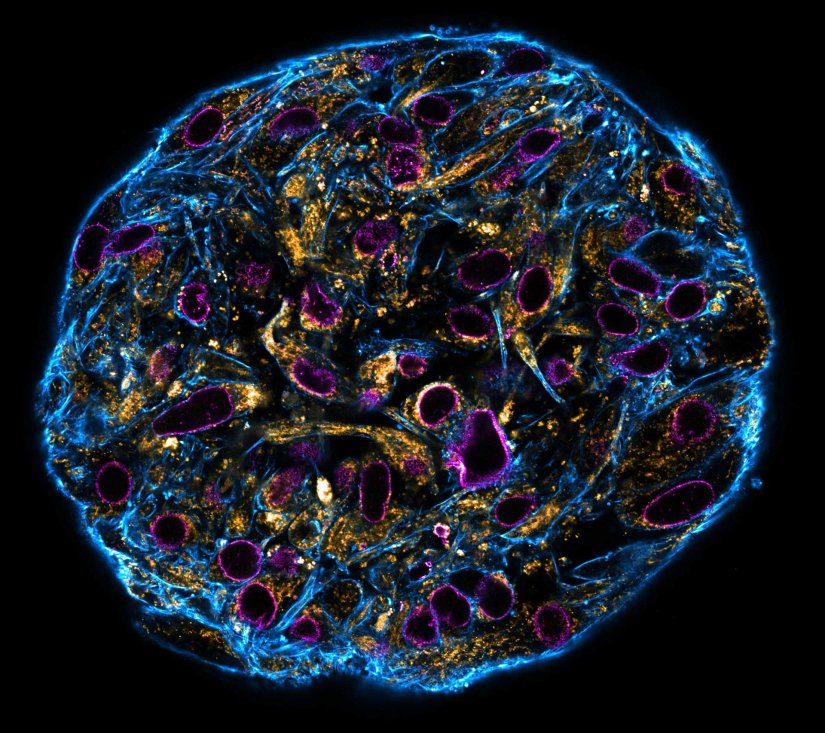
Description
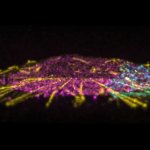
Confocal stitched image of a spheroid stained for nuclear pores (magenta), actin cytoskeleton (blue), and mitochondria (orange).
NIH-313 spheroid, fixed with 4% PFA, kindly provided by ibidi and prepared on µ-Slide VI 0.4 µ-Pattern ibiTreat.
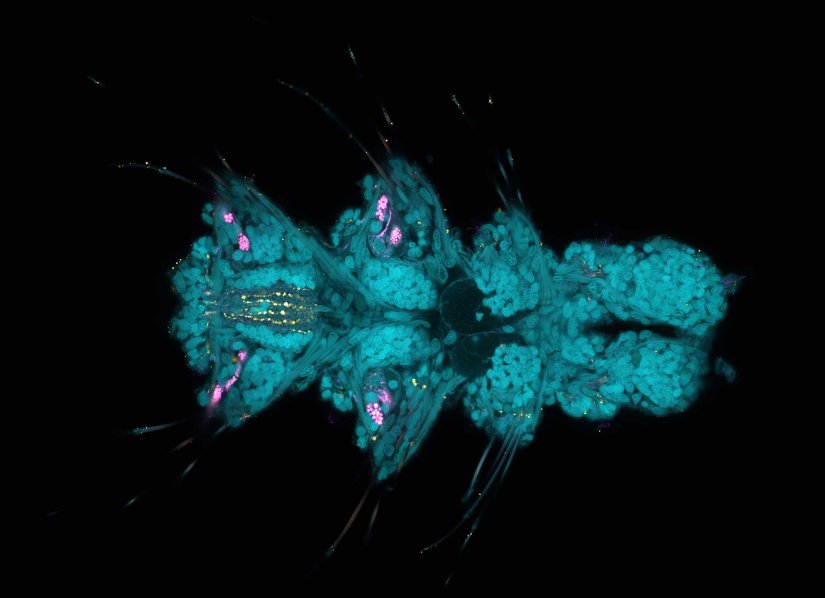
Description
Larva of Platynereis dumerilii acquired in a confocal overview. Nuclei are shown in cyan (DAPI), tubulin in magenta, and serotonin-positive neurons in yellow.
Description
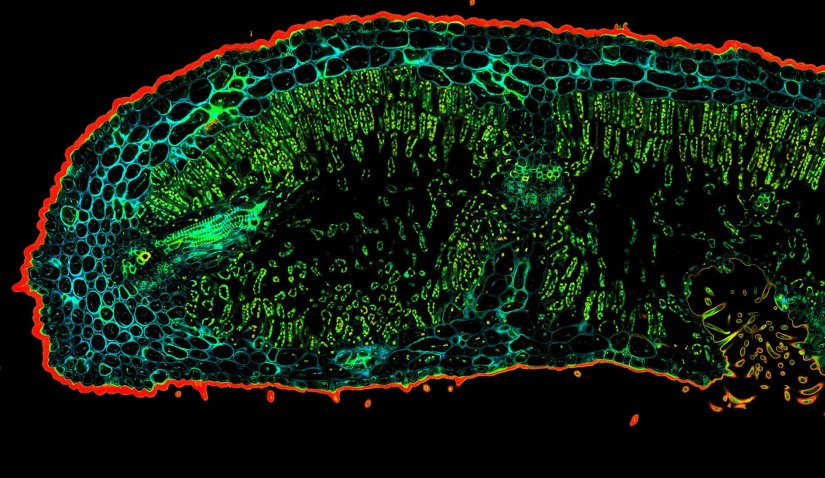
Confocal TIMEBOW acquisition of an Oleander leaf tip showing a range of autofluorescence lifetimes originating from different tissue components.
Description
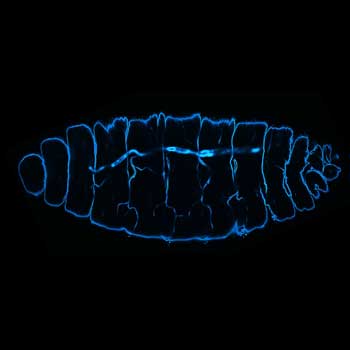
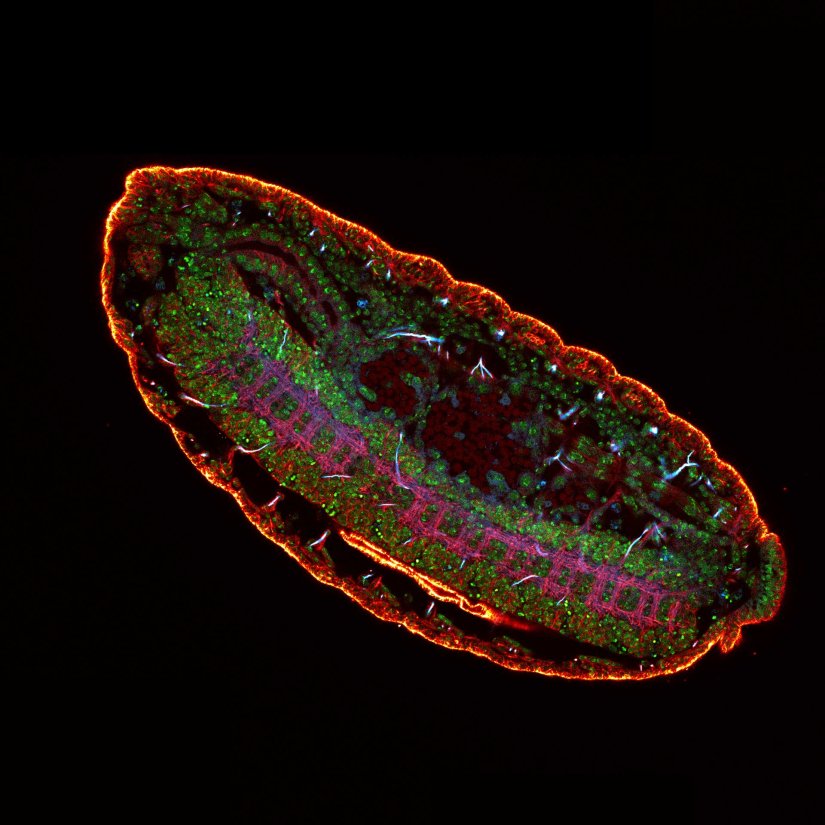
3 color confocal image of a Drosophila embryo stained for chitin (abberior STAR RED, cyan), tubulin (abberior STAR ORANGE, magenta), and DNA (PicoGreen, green).
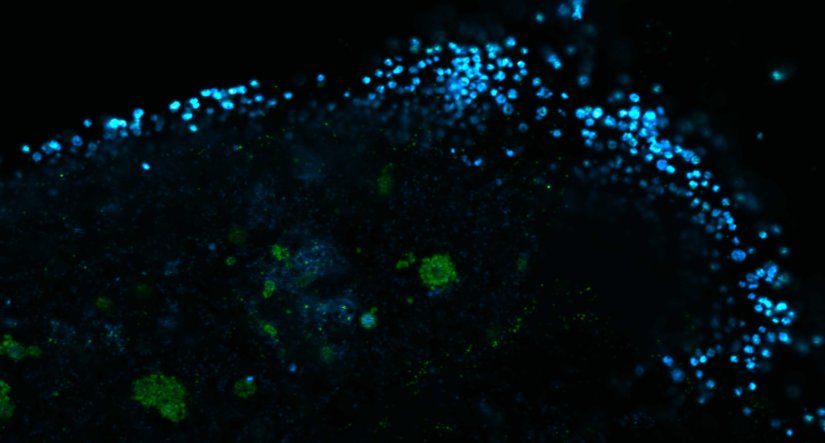
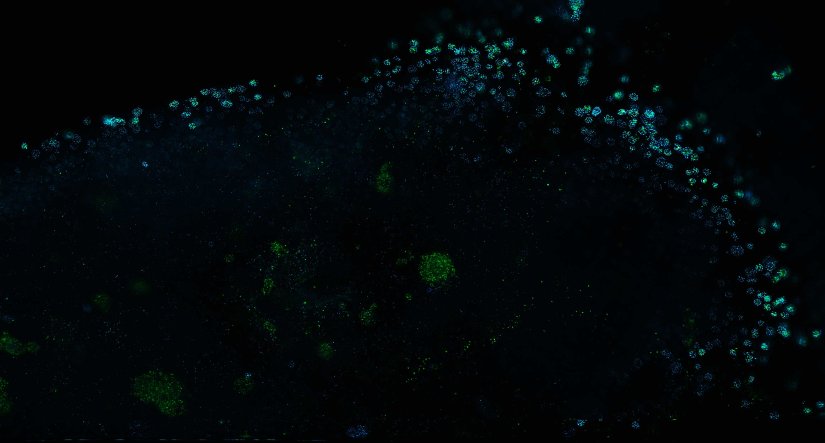
Description
Human cytomegalovirus particles in infected human foreskin fibroblasts, labeled for a viral glycoprotein (abberior STAR RED, blue) and a tegument protein (abberior STAR 580, green). The sample was 4x gel-expanded and imaged with STED.
Description
From old to new with EVERGREEN and STEDYCON! Upgrade your widefield microscope to a confocal and STED system – DFG funding available.

Description
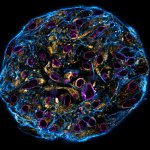
PFA-fixed mammalian cell stained by using Smart Secondaries® from NanoTag Biotechnologies. Stained structures are intermediate filaments (cyan, abberior STAR 460L), mitochondria (green, abberior STAR GREEN), golgi (orange, abberior STAR ORANGE) and nuclear pore complex (magenta, abberior STAR RED).
Description
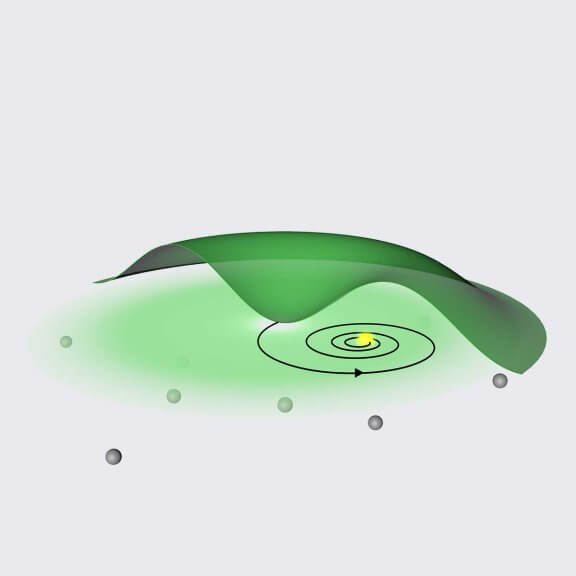
MINFLUX uses information from the emission intensity of a fluorophore to constantly narrow the relative position of the excitation donut and the molecule. This adaptive correction process makes detected photons successively more informative. In contrast, other single-molecule localization techniques gather the exact same information from each detected photon. This way, MINFLUX boosts the summed spatial information gathered from a collection of photons and enables unprecedented resolution without pushing photon numbers or excitation exposure time.
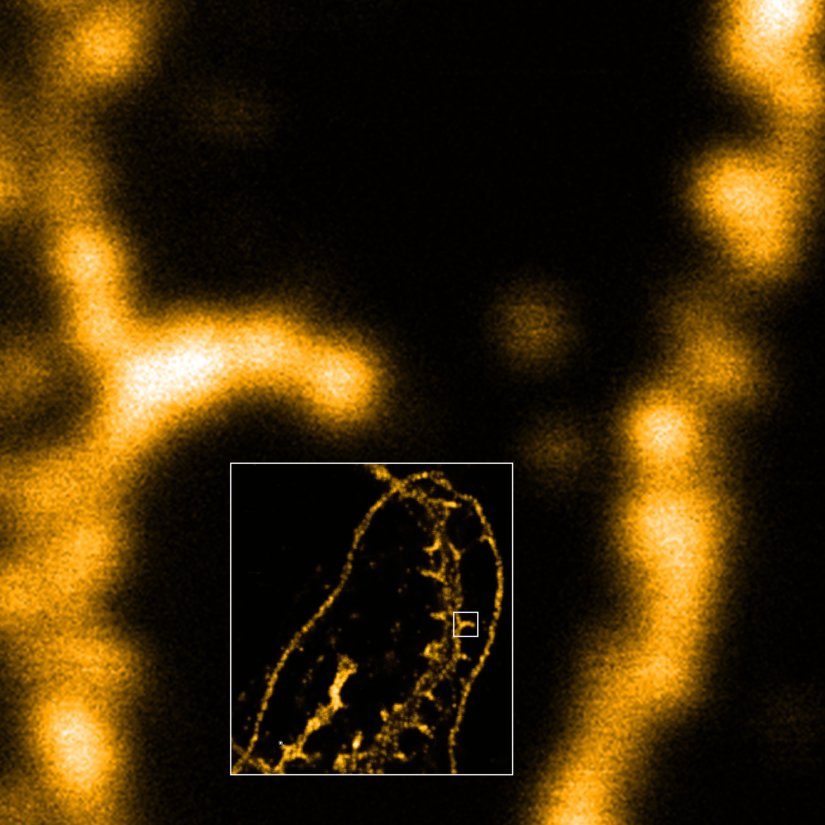
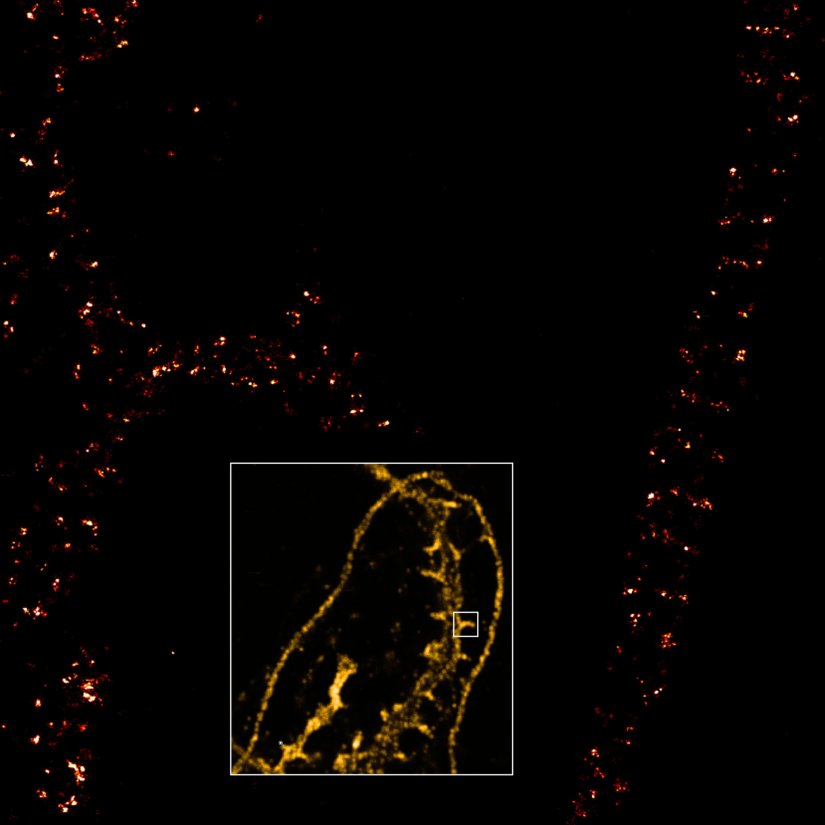
Description
Dendrite with spines of a neuron grown on a coverslip, stained for βII spectrin and imaged in confocal mode and with the MINFLUX module of the MIRAVA POLYSCOPE.
Modules:
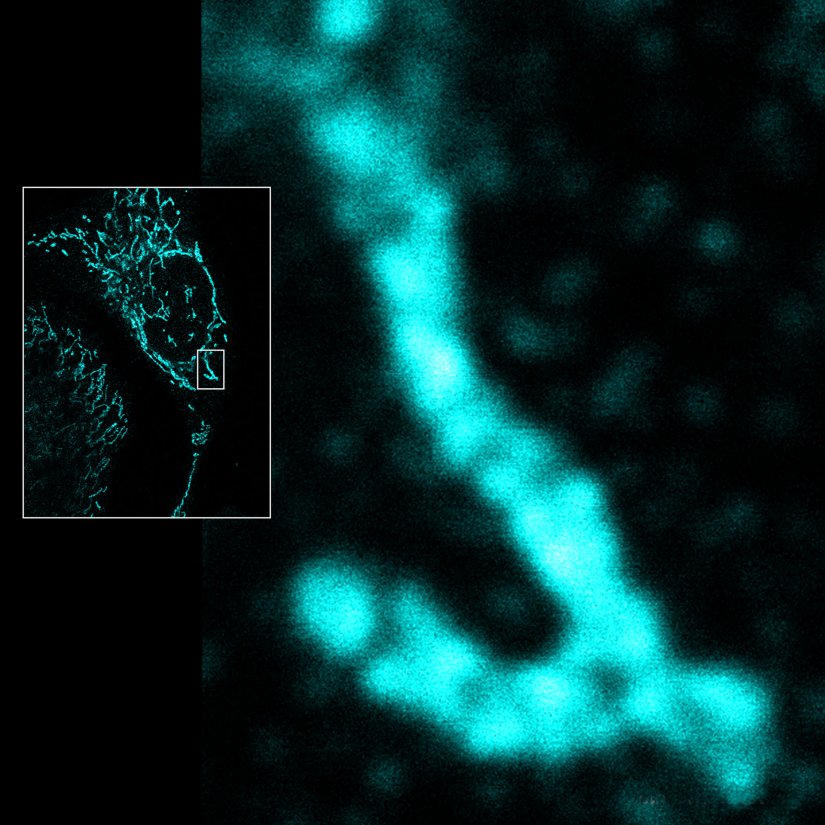
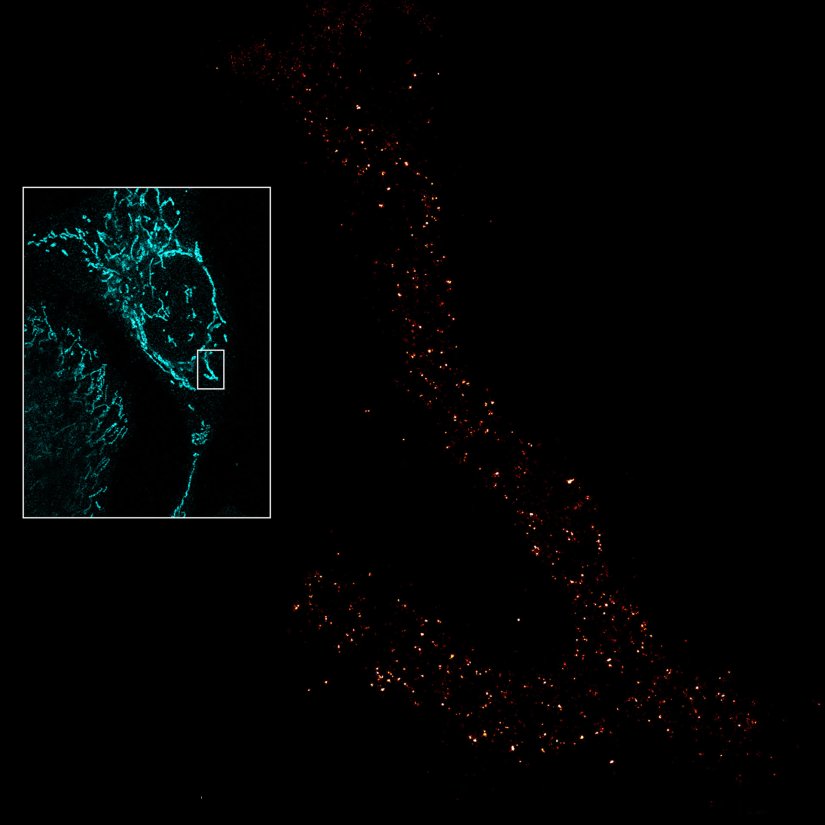
Description
Mitochondria labeled for the outer membrane protein TOMM20, imaged in confocal mode and with the MINFLUX module of the MIRAVA POLYSCOPE.
Modules:
Description
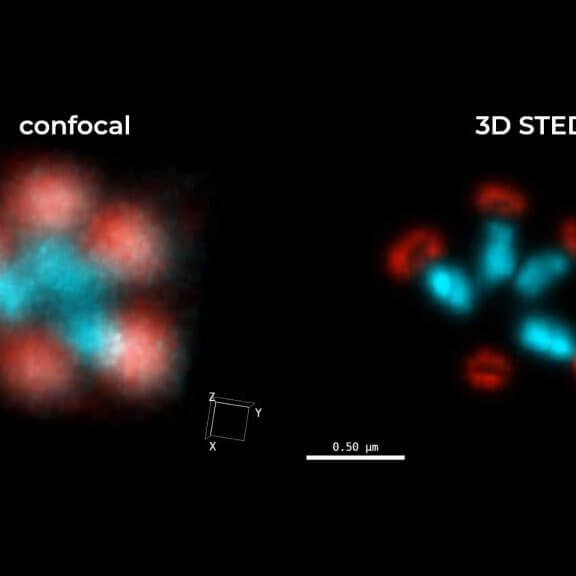
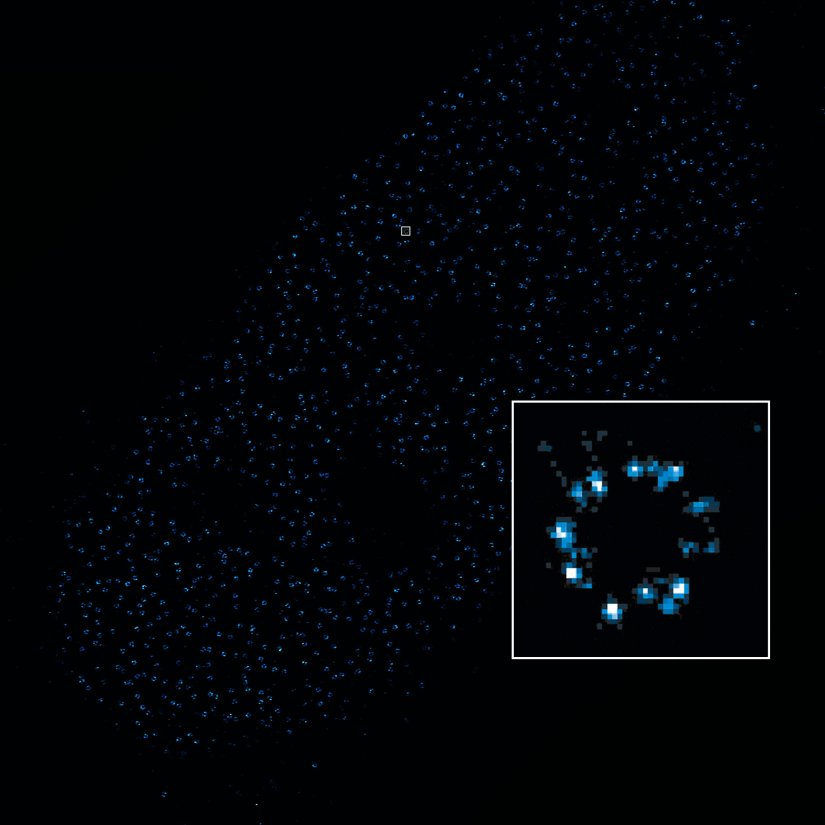
2D MINFLUX image of nuclear pore complex subunits, imaged in fixed mammalian cells expressing GFP-tagged NUP96 stained with abberior DNA-PAINT 660. The molecular resolution of the MINFLUX module of the MIRAVA POLYSCOPE allows visualizing the shape and arrangement of individual subunits of the nuclear pore complex.
Modules:
Description
Description
Confocal image of autofluorescence in a cross-section of the earthworm Lumbricus terrestris. TIMEBOW lifetime imaging detects differences in fluorescence lifetime, which depend on the type of autofluorescent molecule and its nano-environment, and visualizes them in distinct colors.
Modules:
Description
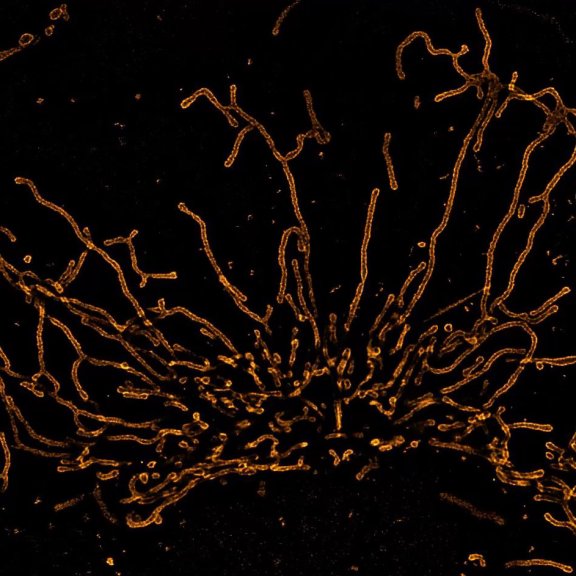
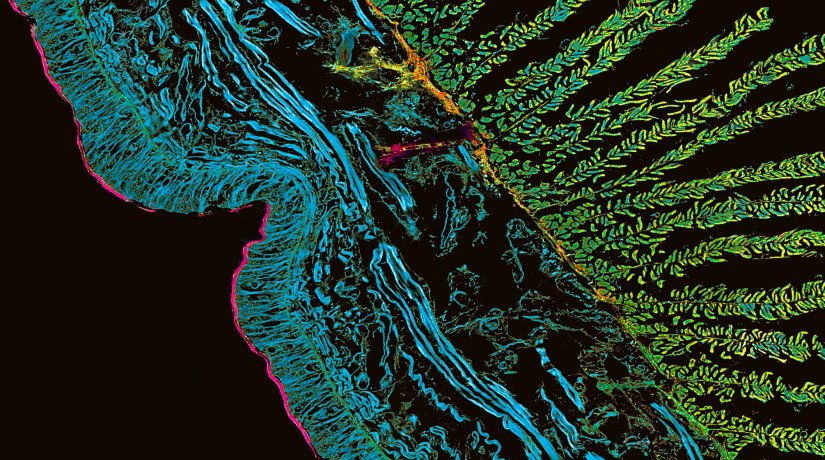
RAYSHAPE aberration correction allows you to look deep into complex samples without losing resolution or brightness due to aberrations. Deep tissue volume of Drosophila trachea showing chitin (autofluorescence) and actin (abberior STAR RED) deconvolved with TRUESHARP.
Modules:
Description
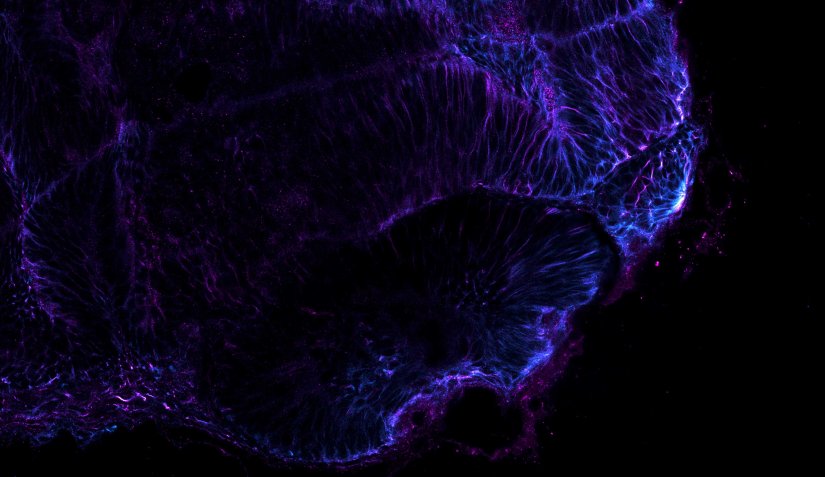
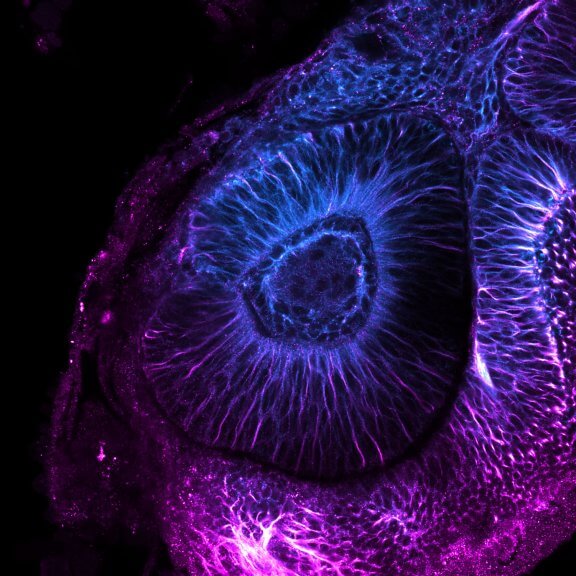
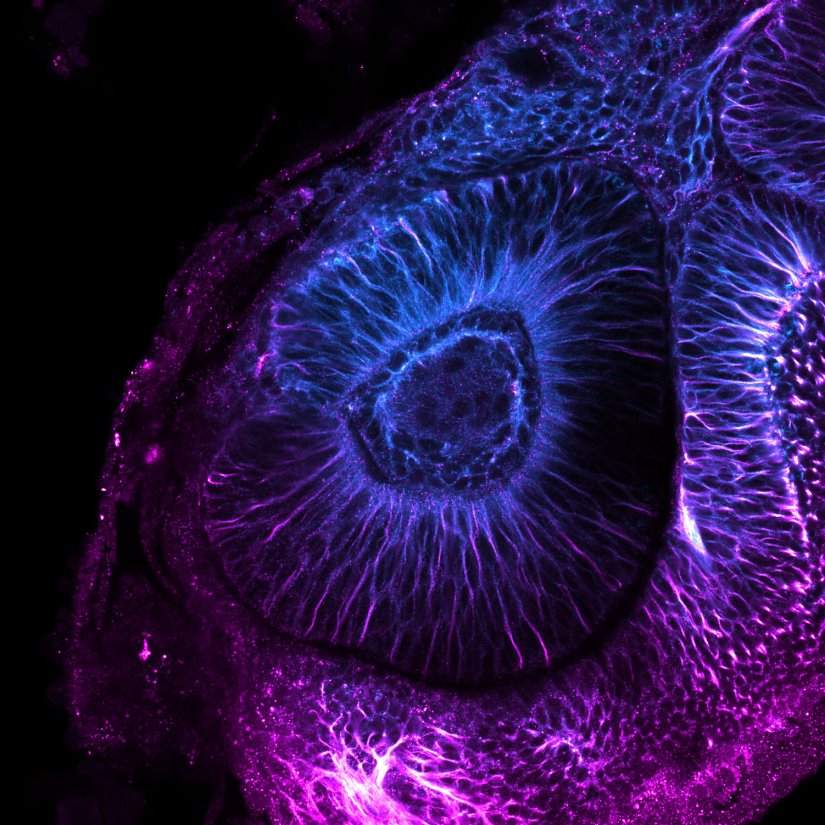
Confocal image of a developing zebrafish eye showing tubulin stained with abberior STAR RED and abberior STAR ORANGE.
Image courtesy of Graziamaria Paradisi, Marco Tartaglia, Antonella Lauri, Ospedale Pediatrico Bambino Gesù, Rome, Italy
Modules:

Description
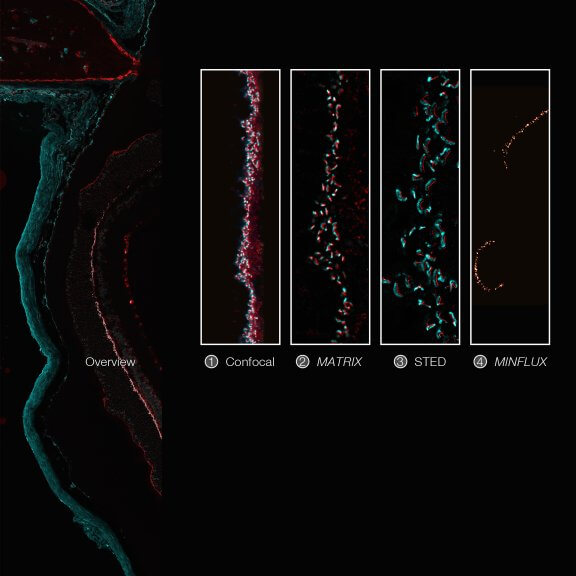
Imaging across scales from diffraction-limited to molecular resolution: ribbon synapses in fixed mouse retina tissue stained for VAMP1 (abberior STAR RED) and CtBP2 (abberior STAR ORANGE) (confocal, MATRIX, STED) or for Bassoon (MINFLUX).
Sample courtesy: Arlene Hirano, PhD, University of California Los Angeles, Los Angeles, CA, USA (confocal, MATRIX, STED), and by Dr. Chad Grabner and Prof. Dr. Tobias Moser, Max Planck Institute for Multidisciplinary Sciences, Göttingen, Germany (MINFLUX).
Modules:
Description
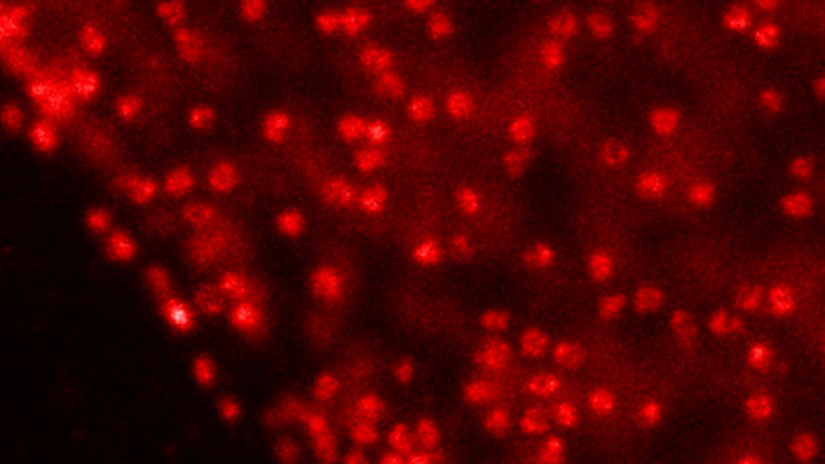
Video of mitochondria in living U2OS cells imaged for 1.5 hours with STED and deconvolved with TRUESHARP image boosting. Mitochondria were visualized with abberior LIVE SiR HaloX staining the outer mitochondrial membrane marker TOMM20. FLEXPOSURE adaptive illumination was used to reduce the light burden on the sample.
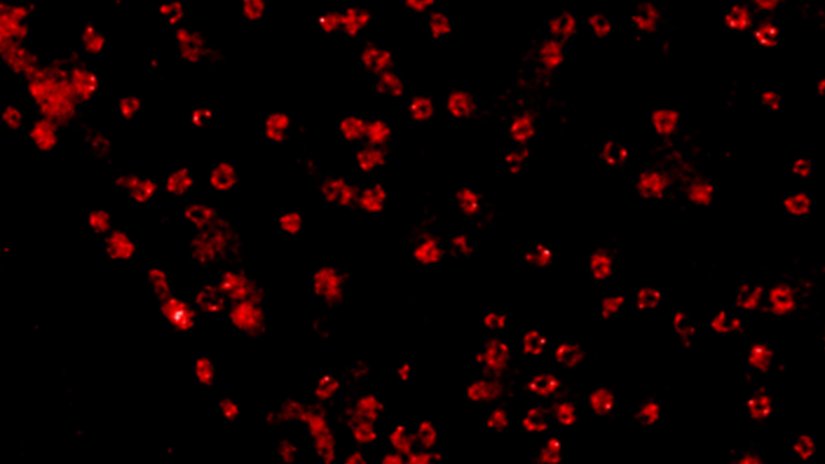
Description
A confocal image compared to a MATRIX + TRUESHARP STED image of nuclear pore complexes in mammalian cells stained for NUP96 with abberior STAR RED.
Modules:
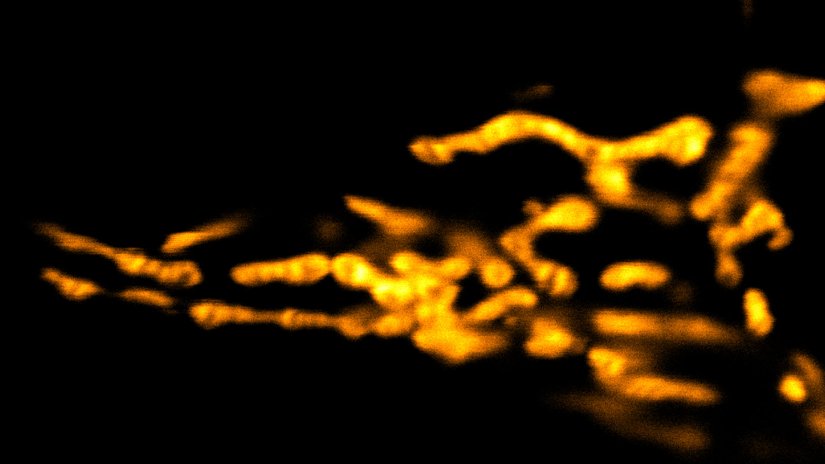
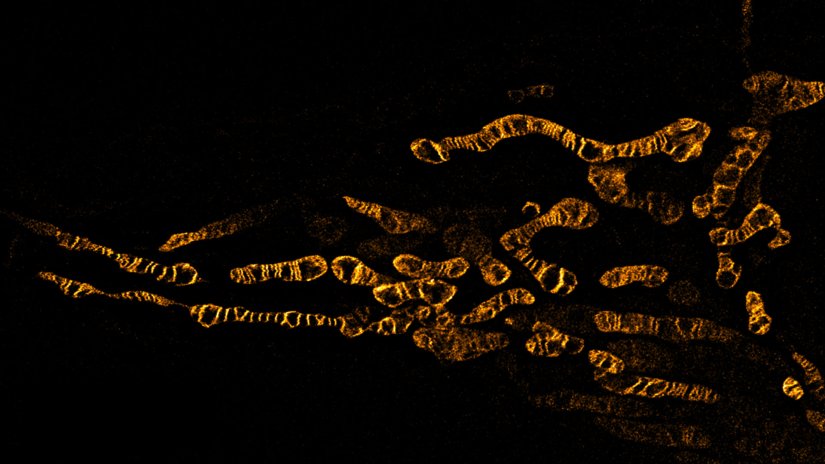
Description
Living HeLa cells stained with the mitochondrial membrane marker abberior LIVE ORANGE mito, visualizing both outer and inner membranes. Confocal and STED images where deconvolved with TRUESHARP image boosting.
Modules:
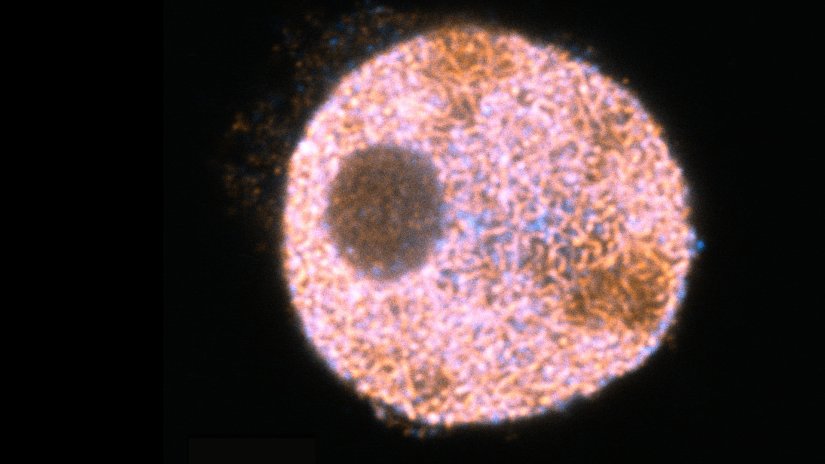
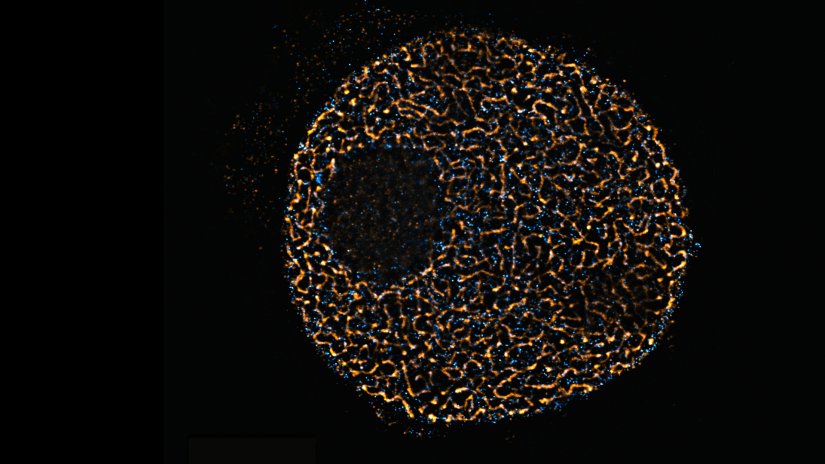
Description
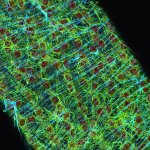
Confocal and STED image of a meiotic cell of Chinese spring wheat stained for two synaptonemal complex components.
Sample courtesy: Sepsi Adél, HUN-REN, Centre for Agricultural Research, Martonvásár, Hungary.
Modules:
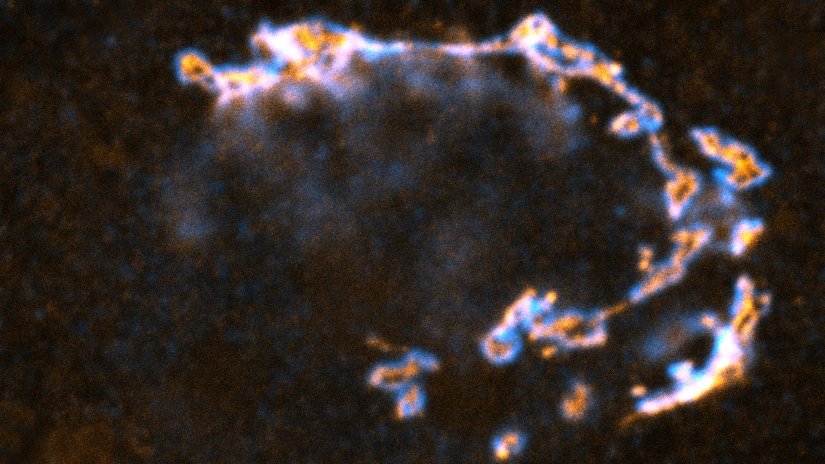

Description
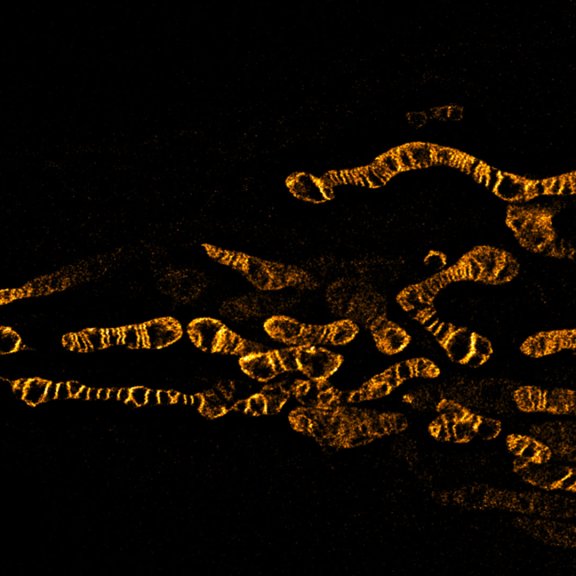
STED image of the golgi apparatus in a fixed mammalian cell, recorded with MATRIX array detection and deconvolved with TRUESHARP image boosting. Stained are the golgi proteins GM130 (cyan, abberior STAR RED) and giantin (orange, abberior STAR ORANGE).
Modules:
Description
Two-color 3D MINFLUX movie revealing an inner and outer mitochondrial membrane marker. Cultured mammalian cells labeled with indirect immunofluorescence using JIR AffiniPure-VHH Fragment antibodies (secondary nanobodies) coupled to abberior FLUX 640 (orange) and FLUX 680 (cyan). MINFLUX enables the visualization and separation of both structures.
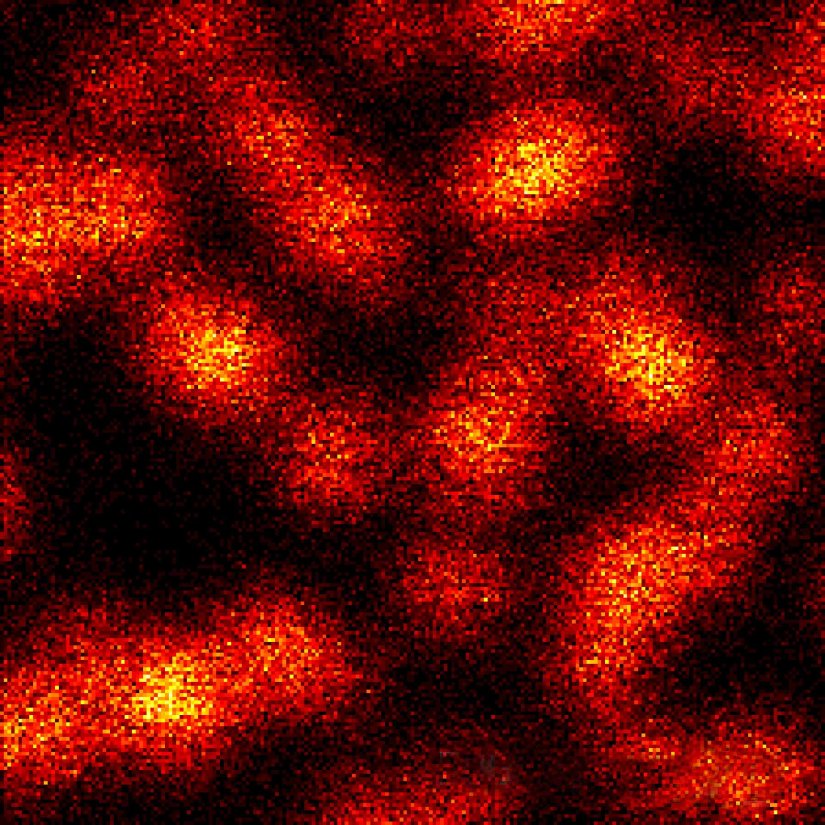

Description
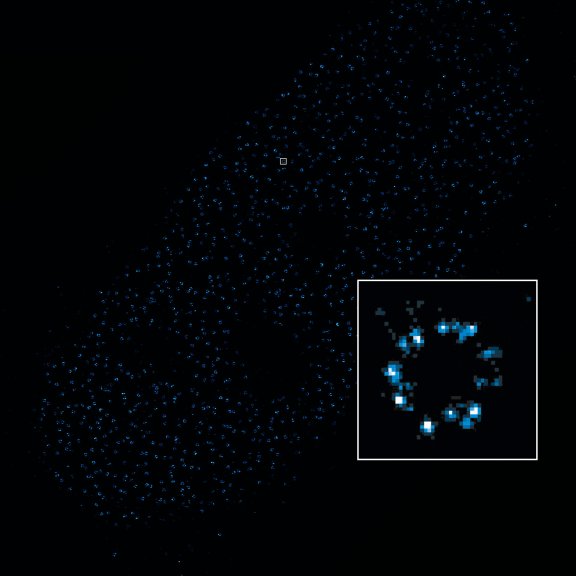
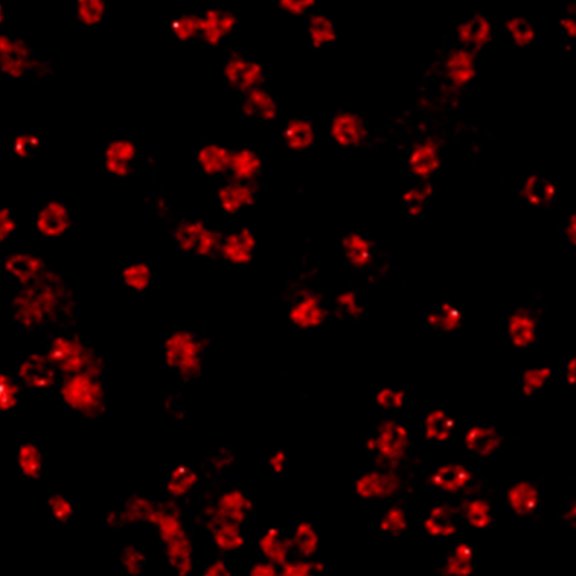
2D MINFLUX nanoscopy of the nuclear pore complex subunits, labeled with abberior FLUX 647 conjugated to JIR AffiniPure-VHH Fragment antibodies (secondary nanobodies). In contrast to confocal microscopy, 2D MINFLUX allows visualization of the shape and arrangement of individual nuclear pore complex subunits. Here, we reach localization precisions of ~ 2 nm in raw localization data.
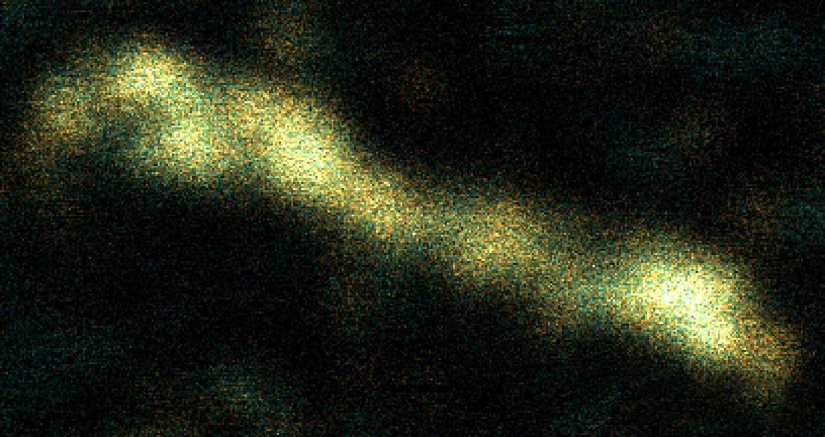

Description
Two-color 3D MINFLUX revealing an inner and outer mitochondrial membrane marker. Cultured mammalian cells labeled with indirect immunofluorescence using JIR AffiniPure-VHH Fragment antibodies (secondary nanobodies) coupled to abberior FLUX 640 (orange) and FLUX 680 (cyan). MINFLUX enables the visualization and separation of both structures.
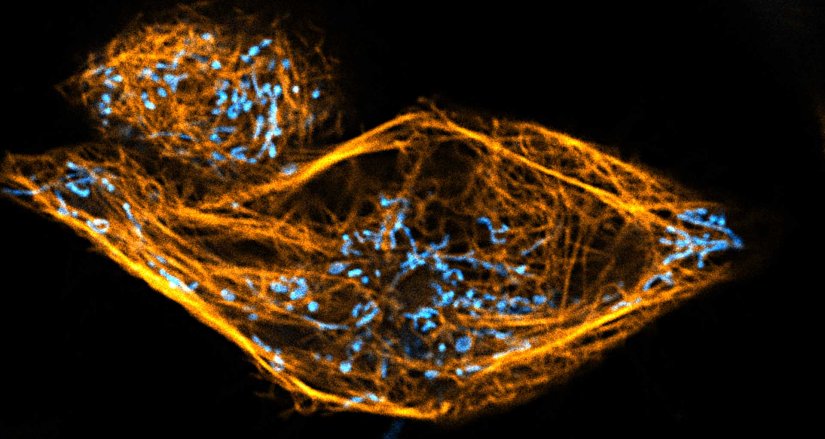
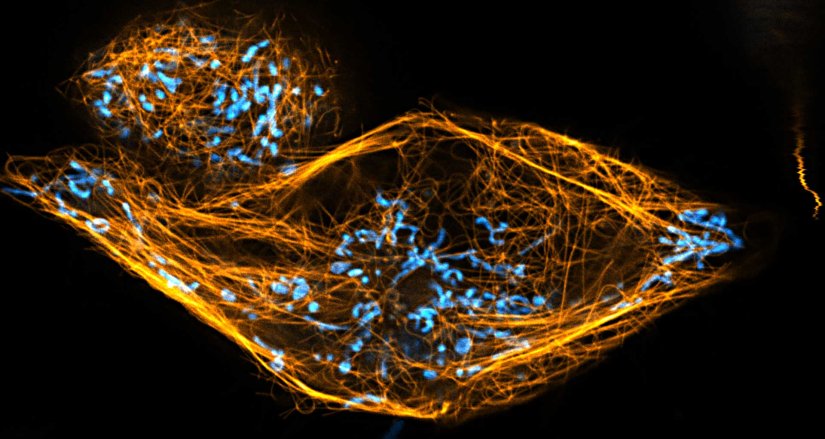
Description
Two color live-cell confocal and STED image of a mammalian cell directly labeled with abberior LIVE 510 mito (cyan), and LIVE RED tubulin. This image was acquired with a STEDYCON microscope.
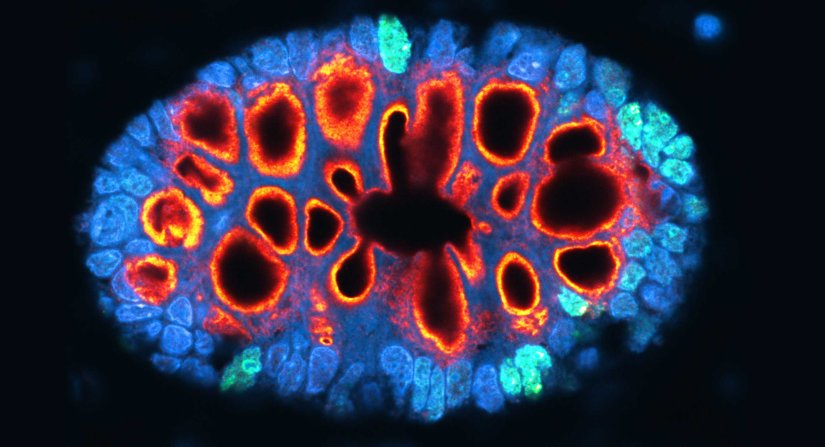
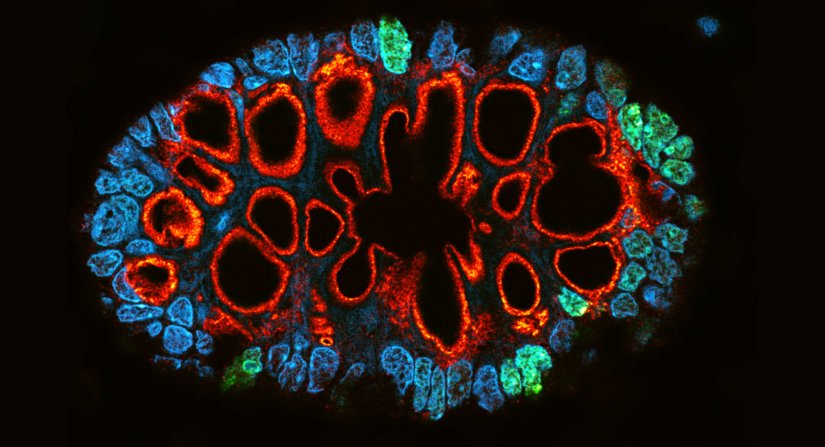
Description
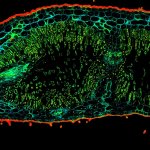
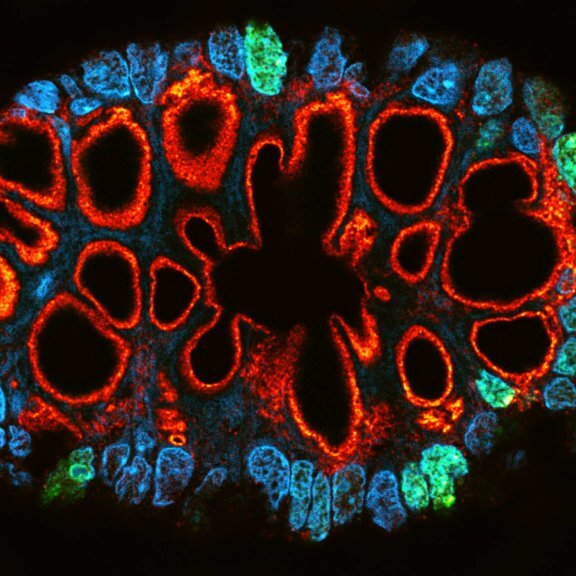
Paraffin section of gut biopsy stained for Ki67 (abberior STAR ORANGE), Muc2 (abberior STAR RED), and DAPI.
Modules:
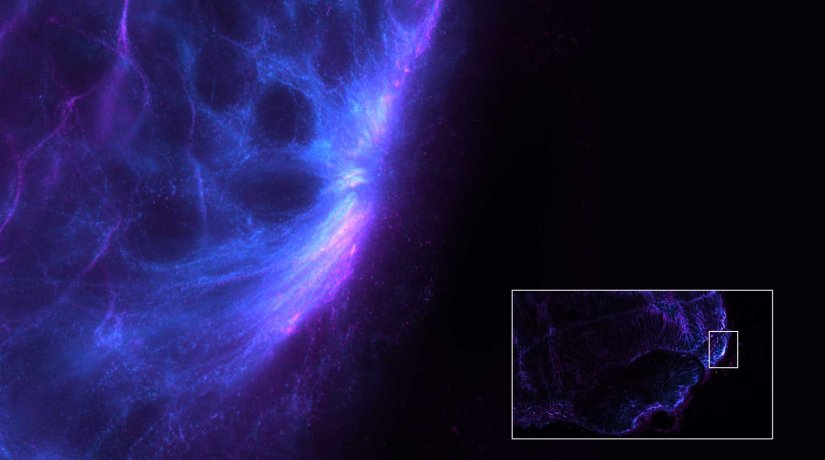
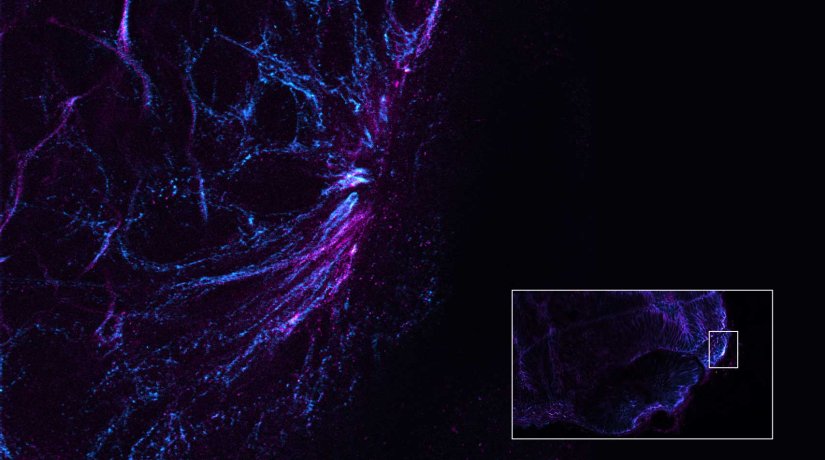
Description
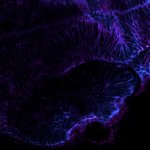
Gain in both signal-to-background ratio and resolution: MATRIX detection dramatically improves a conventional STED image of the zebrafish olfactory epithelium, resulting in a perfect and crystal-clear image revealing even more detail than STED alone can.
Modules:
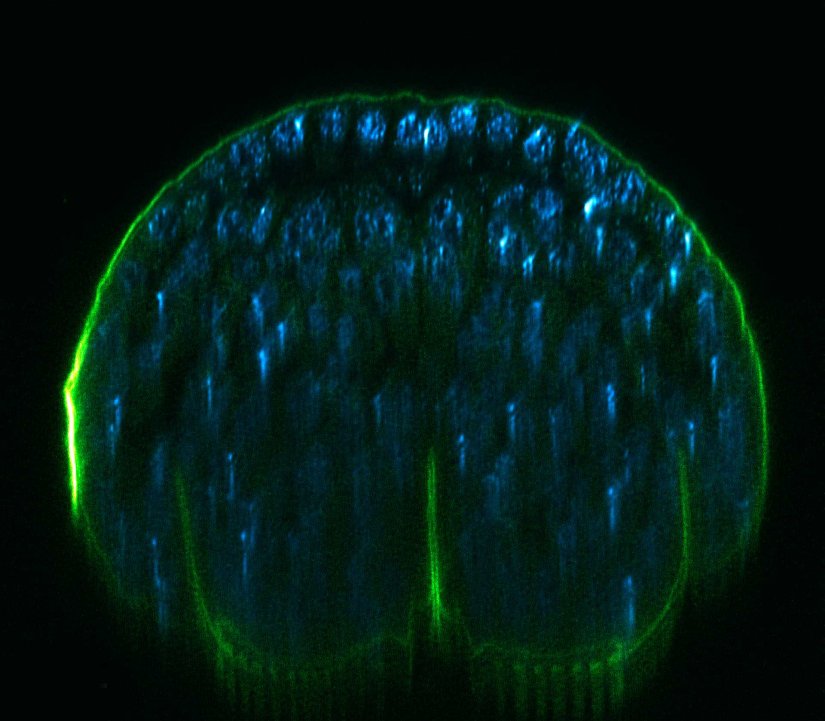
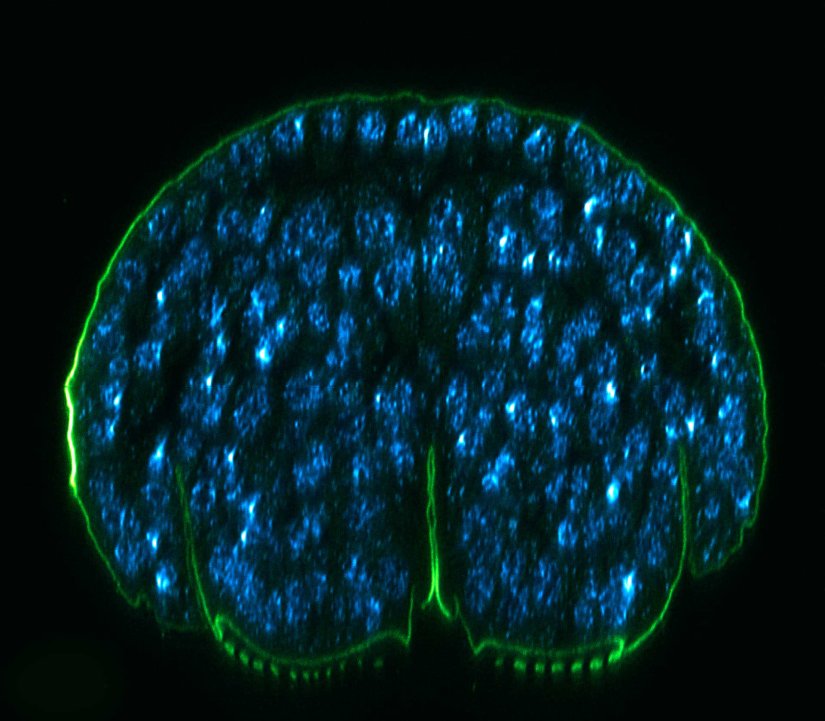
Description
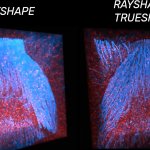
xz section of a stage 17 Drosophila embryo stained for chitin (abberior LIVE 610, green) and DNA (abberior LIVE 550, cyan).
RAYSHAPE preserves resolution and brightness over the whole sample depth of about 200 µm by dynamically redirecting aberrated light to the right places.
In comparison, mechanical optics using a correction collar can only correct a limited z-range of approximately 20 µm
Modules:
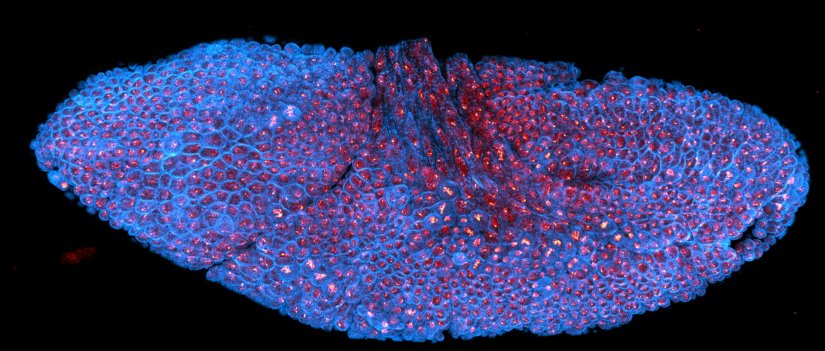
Description
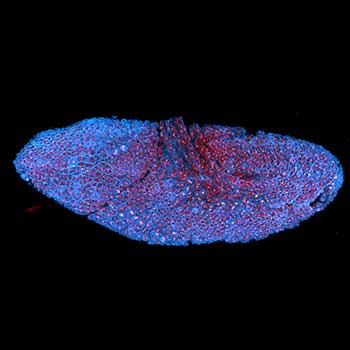
Drosophila stage 12 embryo, imaged with RAYSHAPE, stained for tubulin with abberior STAR RED and for DNA with abberior LIVE 550.
Modules:
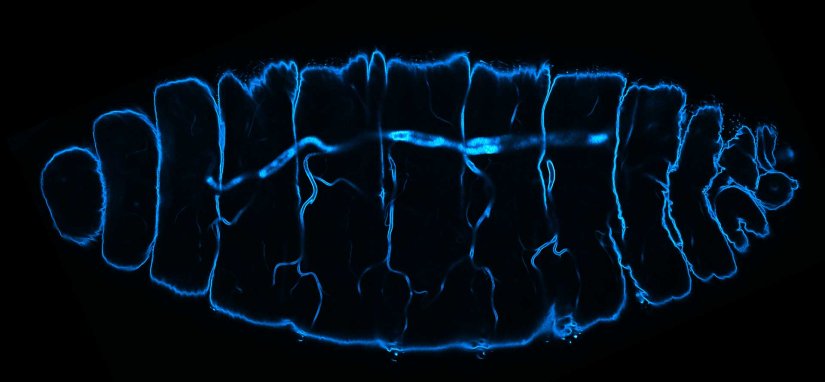
Description
Deep tissue imaging with RAYSHAPE of a stage 17 Drosophila embryo stained for chitin with abberior LIVE 610.
Modules:
Description
Clear and detailed 3D rendering of trachea imaged with RAYSHAPE and MATRIX STED. Chitin was stained with abberior LIVE 610.
Modules:


Description
3D STED xz section of a Drosophila embryo trachea, imaged with RAYSHAPE and without abberation correction, depth 15 µm. Chitin was stained with abberior LIVE 610.
Modules:
Description
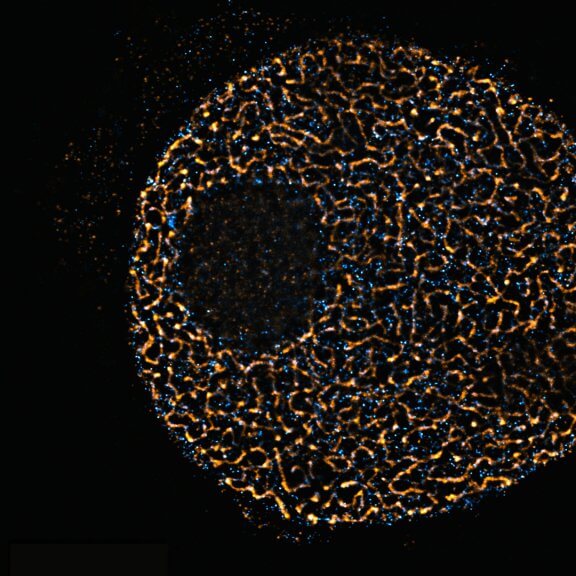
Mammalian cells stained for actin (purple, abberior STAR RED) and vimentin (cyan, abberior LIVE 460L). Differential detection with the MATRIX array detector was used to improve resolution, optical sectioning, and signal-to-background-ratio.
Description
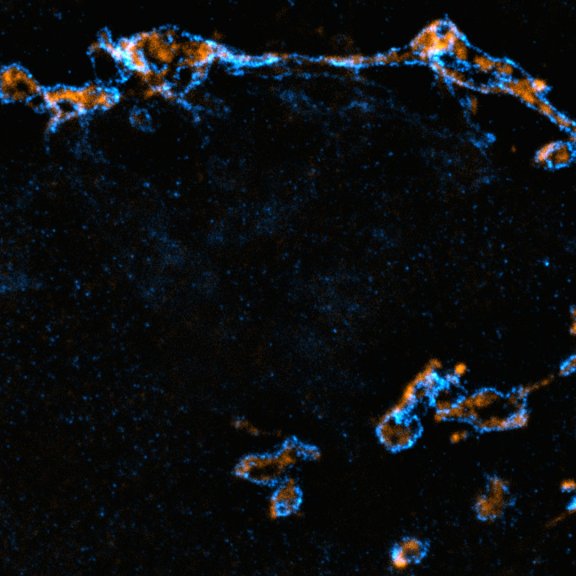
Drosophila ovariole stained for the synaptonemal complex protein C3G (cyan, abberior STAR RED), showing how the complex is formed and disassembled during oocyte development. DNA was stained with DAPI (purple), spectrin with abberior STAR ORANGE (green).
Description
The protozoan Trypanosoma, fixed and three-fold expanded, was stained for tubulin, imaged with STED, and deconvolved with TRUESHARP image boosting. The combination of expansion and STED provides a resolution at which the four tubulin fibers of the flagellum are clearly visible. Scale bar refers to expanded specimen.
Sample courtesy: Mélanie Bonhivers, Laboratoire de Microbiologie Fondamentale et Pathogénicité, CNRS / Université Bordeaux, France
Description
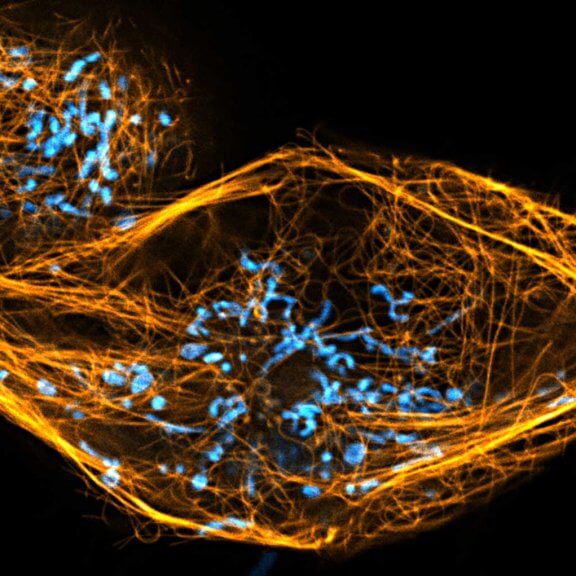
Drosophila egg in the ovary stained for tubulin (green, abberior STAR RED), actin (blue, abberior STAR GREEN), and DNA (red, abberior LIVE 590).
Description
A single 775 nm STED laser was used to visualize this cell’s nuclear pore complex (purple, CF680R), golgi (cyan, abberior STAR RED), and actin (yellow, abberior STAR ORANGE) in superresolution.
Description
A cilium of a fixed RPE-1 cell, gel-expanded and imaged with 3D STED and FLEXPOSURE adaptive illumination. CEP164 was labeled with abberior STAR RED (red) and acetylated tubulin + CCDC41 were labeled with abberior STAR ORANGE (cyan).
Samples courtesy: Dong Kong, PhD & Jadranka Loncarek, PhD. Cancer Innovation Laboratory, NCI/CCR, USA.
Description
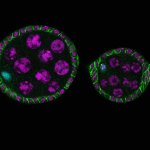
Confocal volume of a Drosophila embryo ventral cord, stained for tubulin (blue) and nuclei (red) and deconvolved with TRUESHARP image boosting.
Description
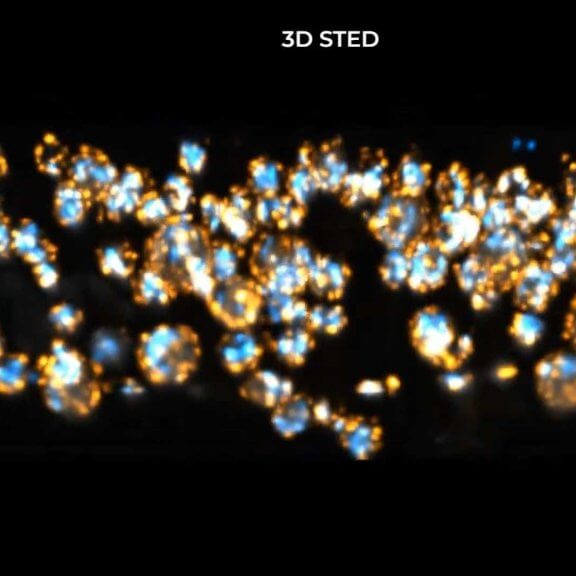
Centriolar rosettes in fixed RPE-1 cells, imaged with 3D STED and FLEXPOSURE adaptive illumination. Labeling: CP110 (abberior STAR RED) and SAS-6 (abberior STAR ORANGE).
Sample courtesy: Catherine Sullenberger & Jadranka Loncarek, Cancer Innovation Laboratory, NCI/CCR, USA
Description
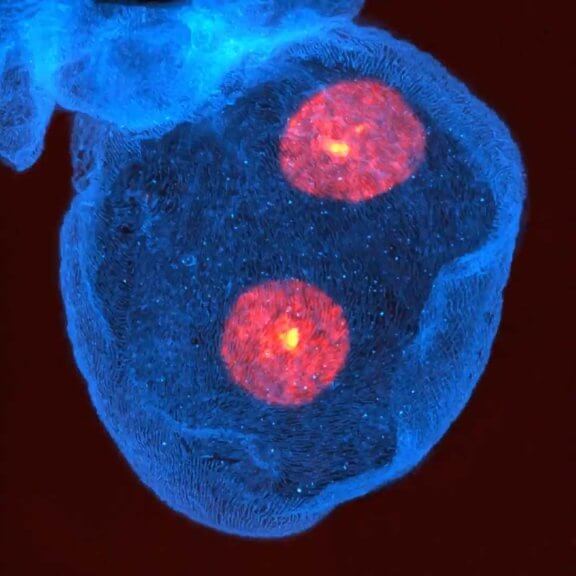
3D STED volume of Drosophila nephrocytes stained for Kirre (cyan, abberior STAR RED), homolog of mammalian Neph1 and a component of the nephrocyte diaphragm, and DAPI (red). Light burden during scan was reduced with FLEXPOSURE adaptive illumination.
Sample courtesy: Konrad Lang, University of Freiburg Medical Center, Germany
Description
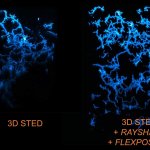
3D STED of microglia and processes labeled for a specific cell surface receptor in neocortical tissue of perfusion-fixed mouse (stained with abberior STAR 635P; unpublished).
FLEXPOSURE adaptive illumination was used to improve signal by reducing light burden and bleaching, RAYSHAPE aberration correction rescued aberration-caused signal loss deep in the sample by adaptive optics.
Sample courtesy: Csaba Cserép and Ádám Dénes, Laboratory of Neuroimmunology, HUN-REN Institute of Experimental Medicine, Budapest, Hungary
Description
Herpes virus particles 4-5x gel-expanded and imaged with confocal and STED, stained for a viral glycoprotein (orange, abberior STAR RED) and tegument protein (cyan, abberior STAR 580).
Description
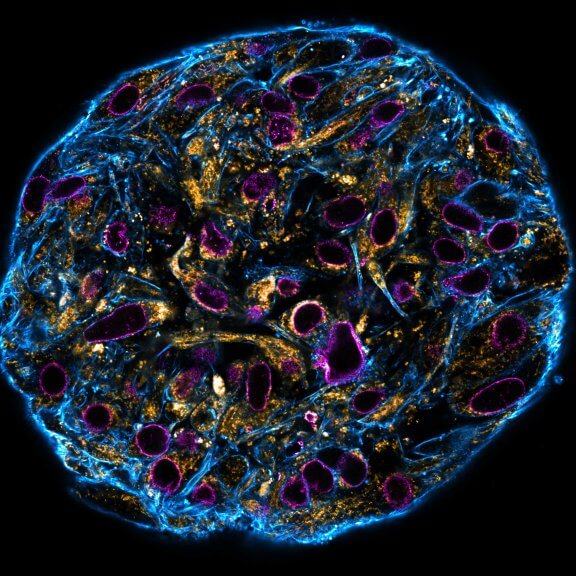
Confocal stitched image of a spheroid stained for nuclear pores (magenta), actin cytoskeleton (blue), and mitochondria (orange).
NIH-313 spheroid, fixed with 4% PFA, kindly provided by ibidi and prepared on µ-Slide VI 0.4 µ-Pattern ibiTreat.
Description
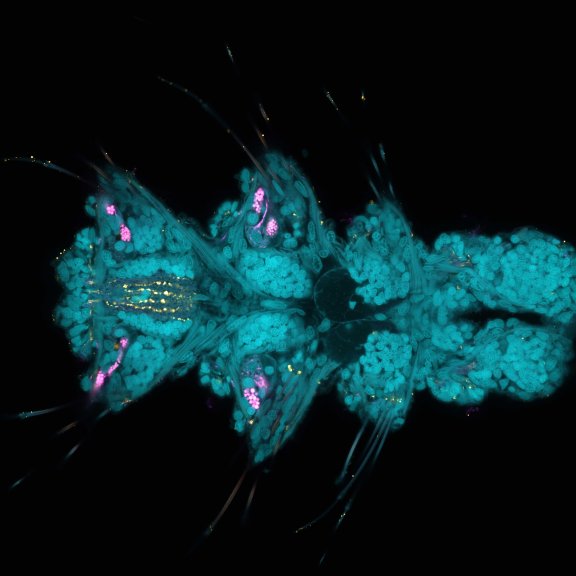
Larva of Platynereis dumerilii acquired in a confocal overview. Nuclei are shown in cyan (DAPI), tubulin in magenta, and serotonin-positive neurons in yellow.
Description
Confocal TIMEBOW acquisition of an Oleander leaf tip showing a range of autofluorescence lifetimes originating from different tissue components.
Description
3 color confocal image of a Drosophila embryo stained for chitin (abberior STAR RED, cyan), tubulin (abberior STAR ORANGE, magenta), and DNA (PicoGreen, green).
Description
Human cytomegalovirus particles in infected human foreskin fibroblasts, labeled for a viral glycoprotein (abberior STAR RED, blue) and a tegument protein (abberior STAR 580, green). The sample was 4x gel-expanded and imaged with STED.
Description
From old to new with EVERGREEN and STEDYCON! Upgrade your widefield microscope to a confocal and STED system – DFG funding available.
Description
PFA-fixed mammalian cell stained by using Smart Secondaries® from NanoTag Biotechnologies. Stained structures are intermediate filaments (cyan, abberior STAR 460L), mitochondria (green, abberior STAR GREEN), golgi (orange, abberior STAR ORANGE) and nuclear pore complex (magenta, abberior STAR RED).
Description
MINFLUX uses information from the emission intensity of a fluorophore to constantly narrow the relative position of the excitation donut and the molecule. This adaptive correction process makes detected photons successively more informative. In contrast, other single-molecule localization techniques gather the exact same information from each detected photon. This way, MINFLUX boosts the summed spatial information gathered from a collection of photons and enables unprecedented resolution without pushing photon numbers or excitation exposure time.
Description
Dendrite with spines of a neuron grown on a coverslip, stained for βII spectrin and imaged in confocal mode and with the MINFLUX module of the MIRAVA POLYSCOPE.
Modules:
Description
Mitochondria labeled for the outer membrane protein TOMM20, imaged in confocal mode and with the MINFLUX module of the MIRAVA POLYSCOPE.
Modules:
Description
2D MINFLUX image of nuclear pore complex subunits, imaged in fixed mammalian cells expressing GFP-tagged NUP96 stained with abberior DNA-PAINT 660. The molecular resolution of the MINFLUX module of the MIRAVA POLYSCOPE allows visualizing the shape and arrangement of individual subunits of the nuclear pore complex.
Modules:
Description
Description
Confocal image of autofluorescence in a cross-section of the earthworm Lumbricus terrestris. TIMEBOW lifetime imaging detects differences in fluorescence lifetime, which depend on the type of autofluorescent molecule and its nano-environment, and visualizes them in distinct colors.
Modules:
Description
RAYSHAPE aberration correction allows you to look deep into complex samples without losing resolution or brightness due to aberrations. Deep tissue volume of Drosophila trachea showing chitin (autofluorescence) and actin (abberior STAR RED) deconvolved with TRUESHARP.
Modules:
Description
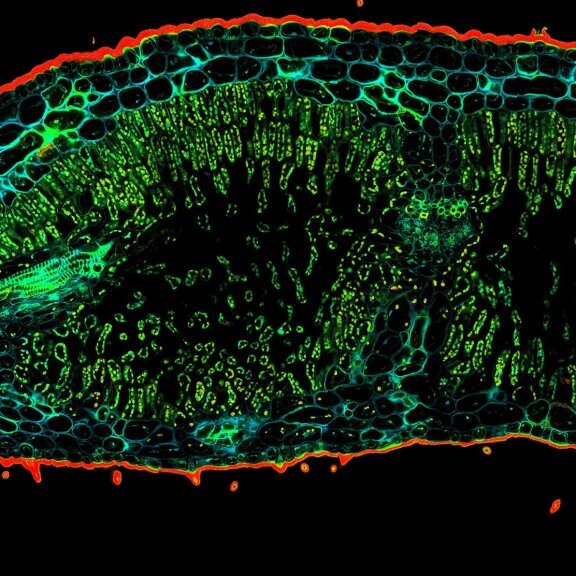
Confocal image of a developing zebrafish eye showing tubulin stained with abberior STAR RED and abberior STAR ORANGE.
Image courtesy of Graziamaria Paradisi, Marco Tartaglia, Antonella Lauri, Ospedale Pediatrico Bambino Gesù, Rome, Italy
Modules:
Description
Imaging across scales from diffraction-limited to molecular resolution: ribbon synapses in fixed mouse retina tissue stained for VAMP1 (abberior STAR RED) and CtBP2 (abberior STAR ORANGE) (confocal, MATRIX, STED) or for Bassoon (MINFLUX).
Sample courtesy: Arlene Hirano, PhD, University of California Los Angeles, Los Angeles, CA, USA (confocal, MATRIX, STED), and by Dr. Chad Grabner and Prof. Dr. Tobias Moser, Max Planck Institute for Multidisciplinary Sciences, Göttingen, Germany (MINFLUX).
Modules:
Description
Video of mitochondria in living U2OS cells imaged for 1.5 hours with STED and deconvolved with TRUESHARP image boosting. Mitochondria were visualized with abberior LIVE SiR HaloX staining the outer mitochondrial membrane marker TOMM20. FLEXPOSURE adaptive illumination was used to reduce the light burden on the sample.
Modules:
Description
A confocal image compared to a MATRIX + TRUESHARP STED image of nuclear pore complexes in mammalian cells stained for NUP96 with abberior STAR RED.
Modules:
Description
Living HeLa cells stained with the mitochondrial membrane marker abberior LIVE ORANGE mito, visualizing both outer and inner membranes. Confocal and STED images where deconvolved with TRUESHARP image boosting.
Modules:
Description
Confocal and STED image of a meiotic cell of Chinese spring wheat stained for two synaptonemal complex components.
Sample courtesy: Sepsi Adél, HUN-REN, Centre for Agricultural Research, Martonvásár, Hungary.
Modules:
Description
STED image of the golgi apparatus in a fixed mammalian cell, recorded with MATRIX array detection and deconvolved with TRUESHARP image boosting. Stained are the golgi proteins GM130 (cyan, abberior STAR RED) and giantin (orange, abberior STAR ORANGE).
Modules:
Description
Two-color 3D MINFLUX movie revealing an inner and outer mitochondrial membrane marker. Cultured mammalian cells labeled with indirect immunofluorescence using JIR AffiniPure-VHH Fragment antibodies (secondary nanobodies) coupled to abberior FLUX 640 (orange) and FLUX 680 (cyan). MINFLUX enables the visualization and separation of both structures.
Description
2D MINFLUX nanoscopy of the nuclear pore complex subunits, labeled with abberior FLUX 647 conjugated to JIR AffiniPure-VHH Fragment antibodies (secondary nanobodies). In contrast to confocal microscopy, 2D MINFLUX allows visualization of the shape and arrangement of individual nuclear pore complex subunits. Here, we reach localization precisions of ~ 2 nm in raw localization data.
Description
Two-color 3D MINFLUX revealing an inner and outer mitochondrial membrane marker. Cultured mammalian cells labeled with indirect immunofluorescence using JIR AffiniPure-VHH Fragment antibodies (secondary nanobodies) coupled to abberior FLUX 640 (orange) and FLUX 680 (cyan). MINFLUX enables the visualization and separation of both structures.
Description
Two color live-cell confocal and STED image of a mammalian cell directly labeled with abberior LIVE 510 mito (cyan), and LIVE RED tubulin. This image was acquired with a STEDYCON microscope.
Description
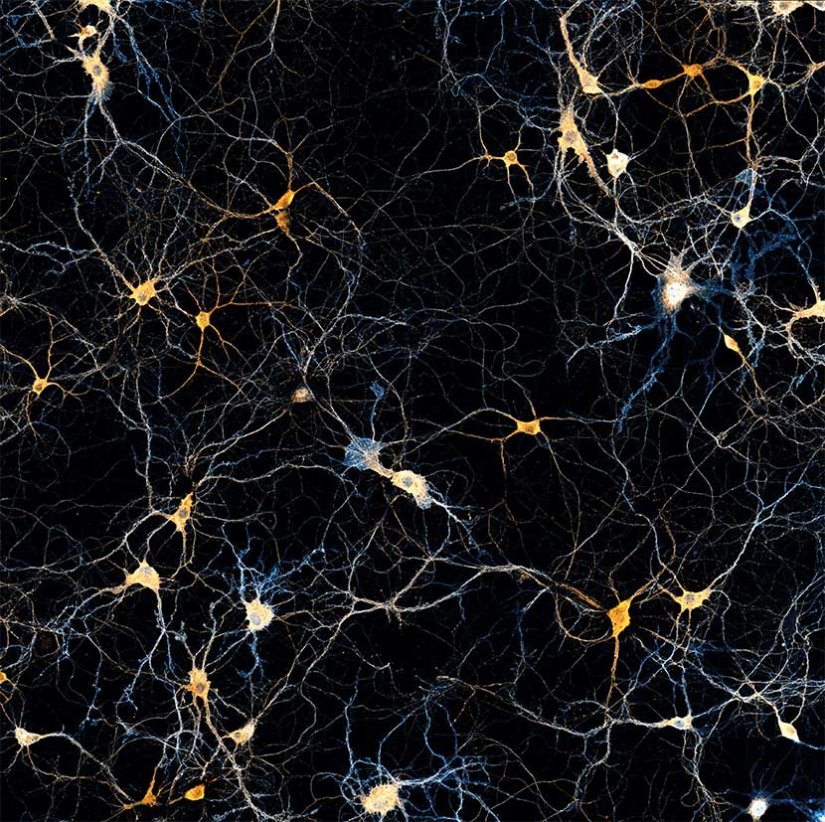
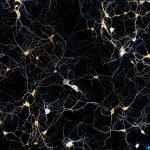
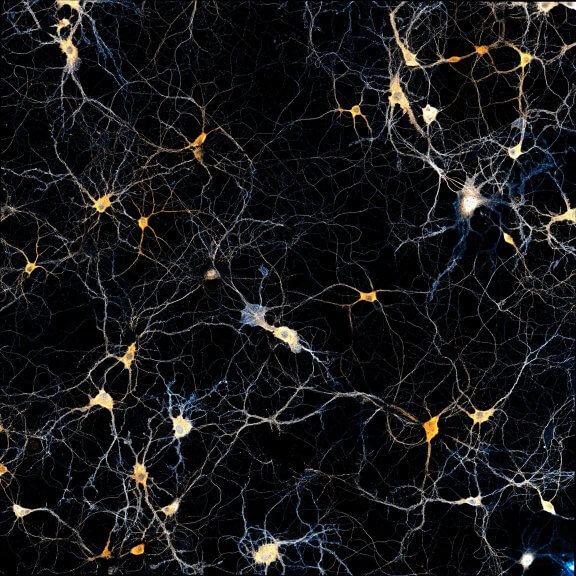
A brain section stained for the neuronal proteins spectrin and adducine.
Description
Paraffin section of gut biopsy stained for Ki67 (abberior STAR ORANGE), Muc2 (abberior STAR RED), and DAPI.
Modules:
Description
Gain in both signal-to-background ratio and resolution: MATRIX detection dramatically improves a conventional STED image of the zebrafish olfactory epithelium, resulting in a perfect and crystal-clear image revealing even more detail than STED alone can.
Modules:
Description
xz section of a stage 17 Drosophila embryo stained for chitin (abberior LIVE 610, green) and DNA (abberior LIVE 550, cyan).
RAYSHAPE preserves resolution and brightness over the whole sample depth of about 200 µm by dynamically redirecting aberrated light to the right places.
In comparison, mechanical optics using a correction collar can only correct a limited z-range of approximately 20 µm
Modules:
Description
Drosophila stage 12 embryo, imaged with RAYSHAPE, stained for tubulin with abberior STAR RED and for DNA with abberior LIVE 550.
Modules:
Description
Deep tissue imaging with RAYSHAPE of a stage 17 Drosophila embryo stained for chitin with abberior LIVE 610.
Modules:
Description
Clear and detailed 3D rendering of trachea imaged with RAYSHAPE and MATRIX STED. Chitin was stained with abberior LIVE 610.
Modules:
Description
3D STED xz section of a Drosophila embryo trachea, imaged with RAYSHAPE and without abberation correction, depth 15 µm. Chitin was stained with abberior LIVE 610.