Sample gallery
Fluorescence imaging, whether at confocal, STED or MINFLUX resolution, guarantees unique insights into the function and structure of life at the molecular level. Besides the scientific information content, some sample portraits provide simply beautiful images. Enjoy browsing our sample gallery.
the fine art of science

Description
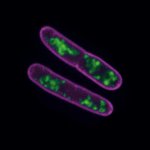
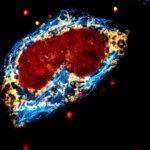
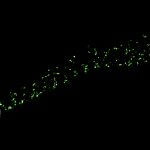
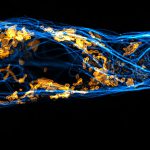
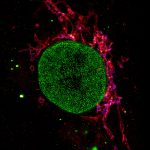
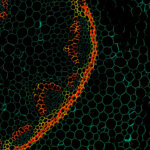
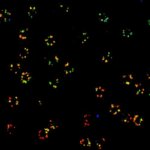
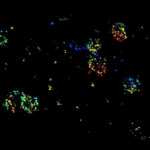
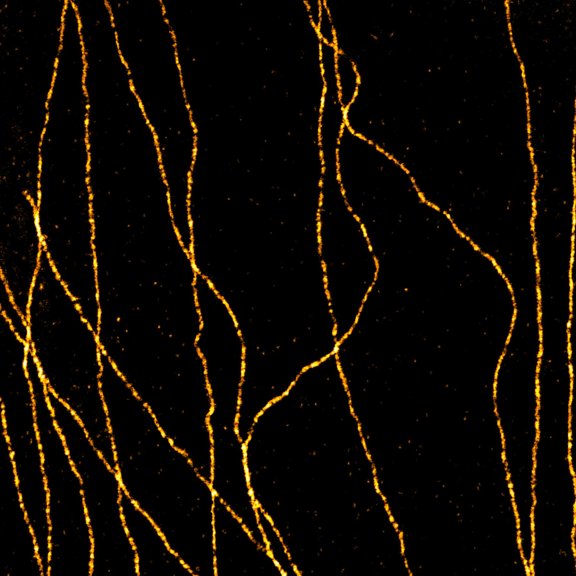
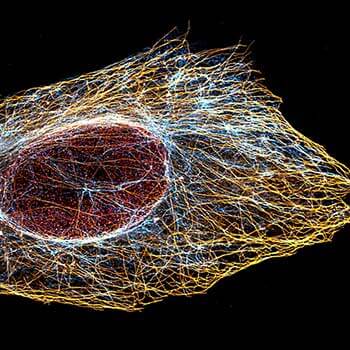
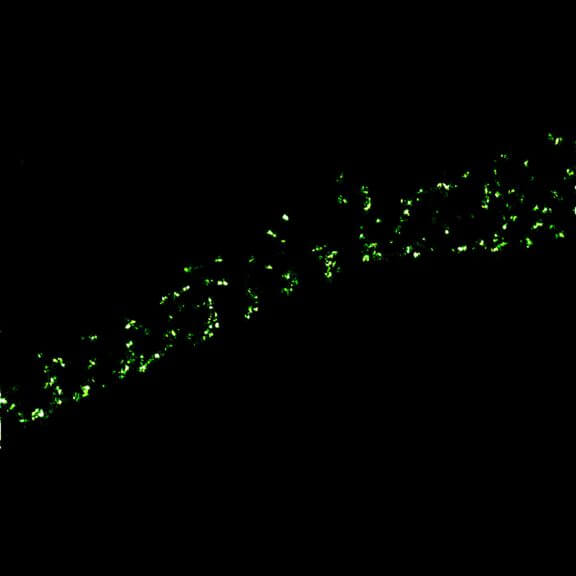
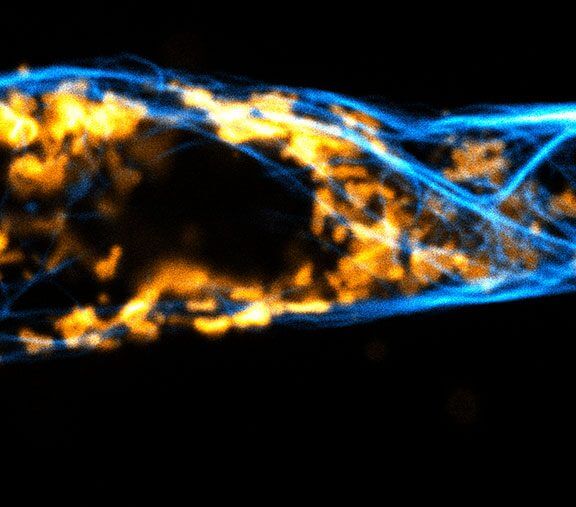
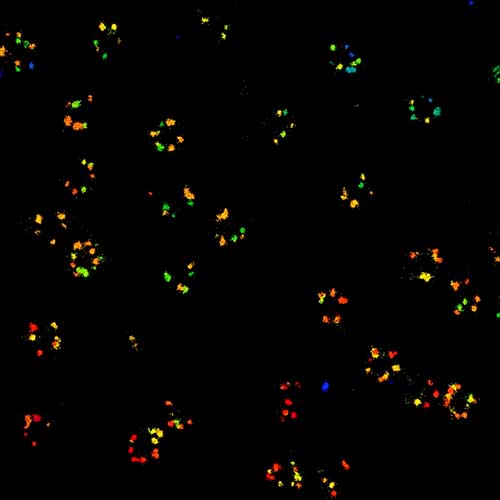
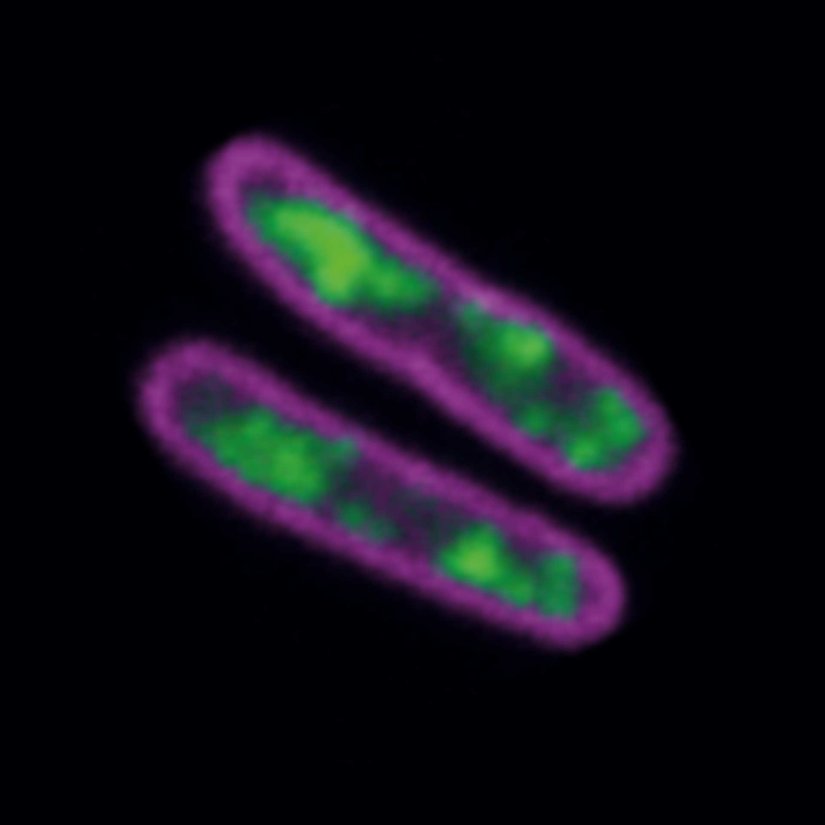
2D MINFLUX nanoscopy of Y. enterocolitica sorting platform protein Halo-YscL.
Sample from Alexander Carsten, Prof. Martin Aepfelbacher, Institute of Medical Microbiology, Virology and Hygiene, University Medical Center Hamburg Eppendorf, Germany
Description
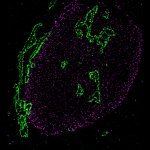
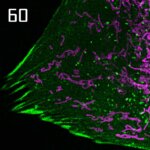
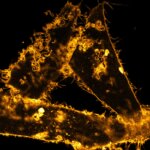
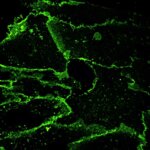
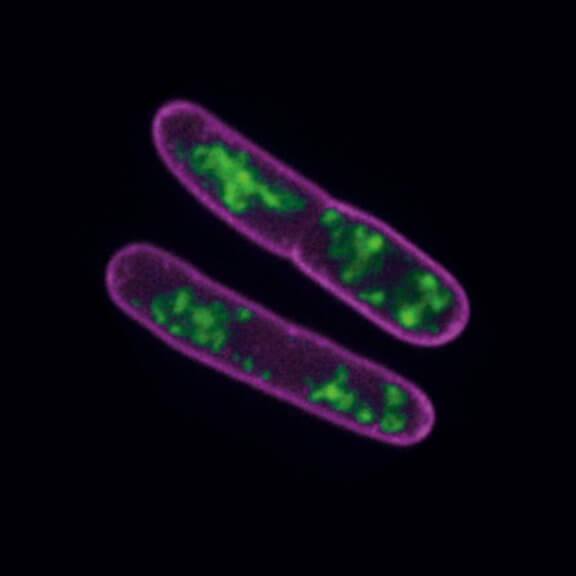
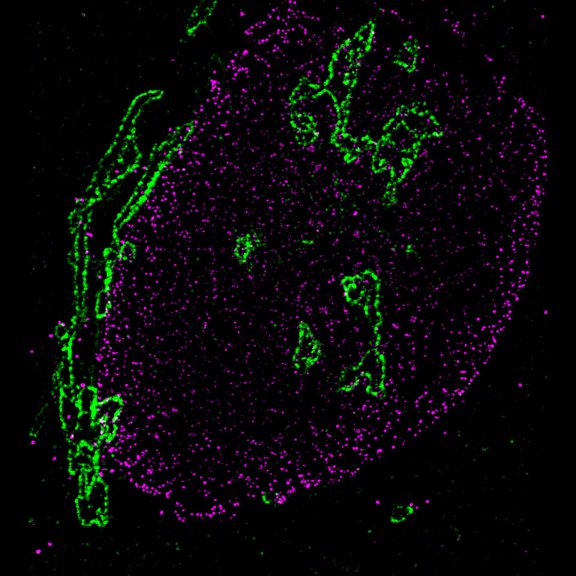
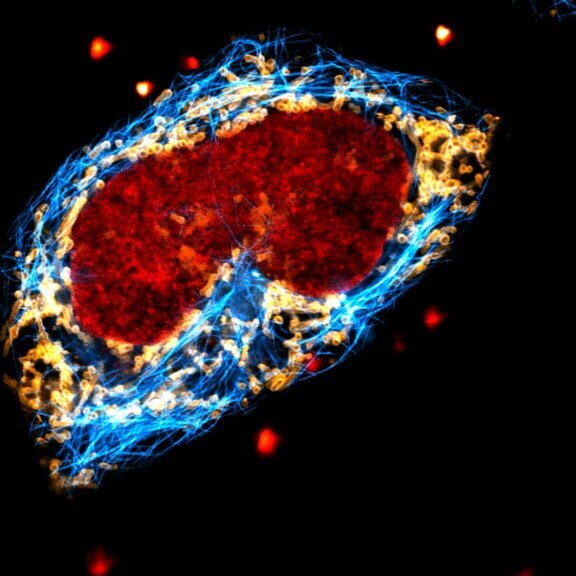
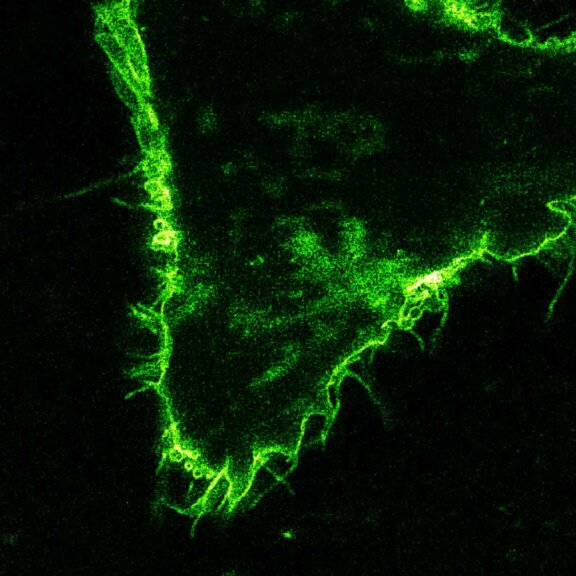
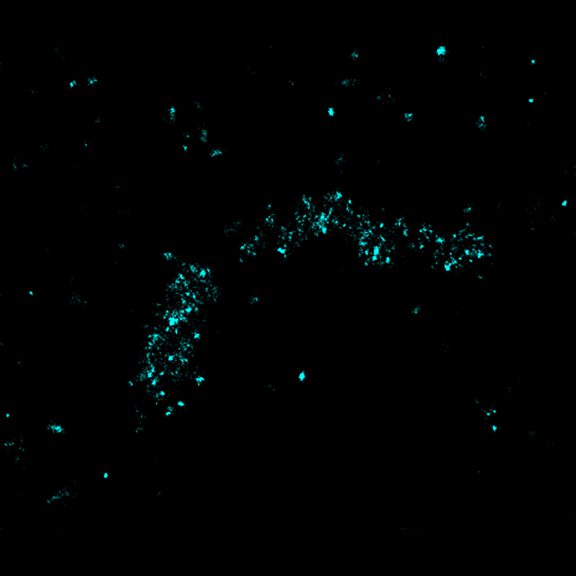
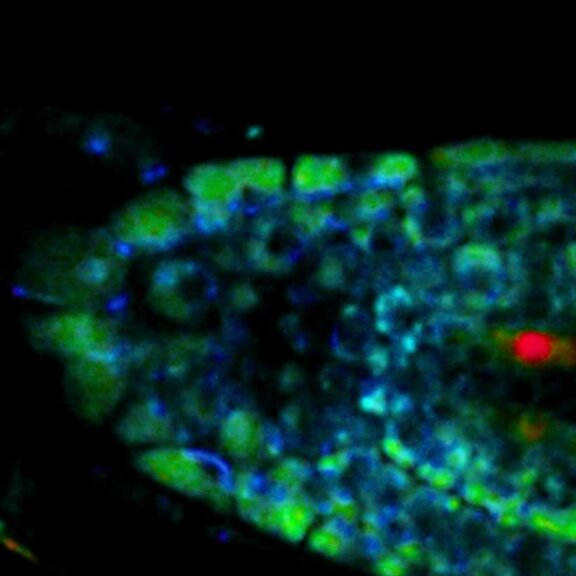
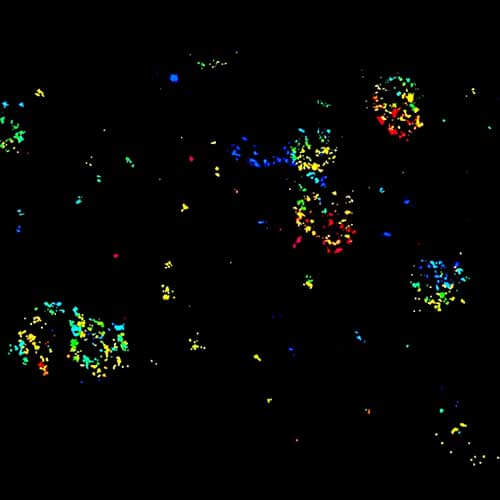
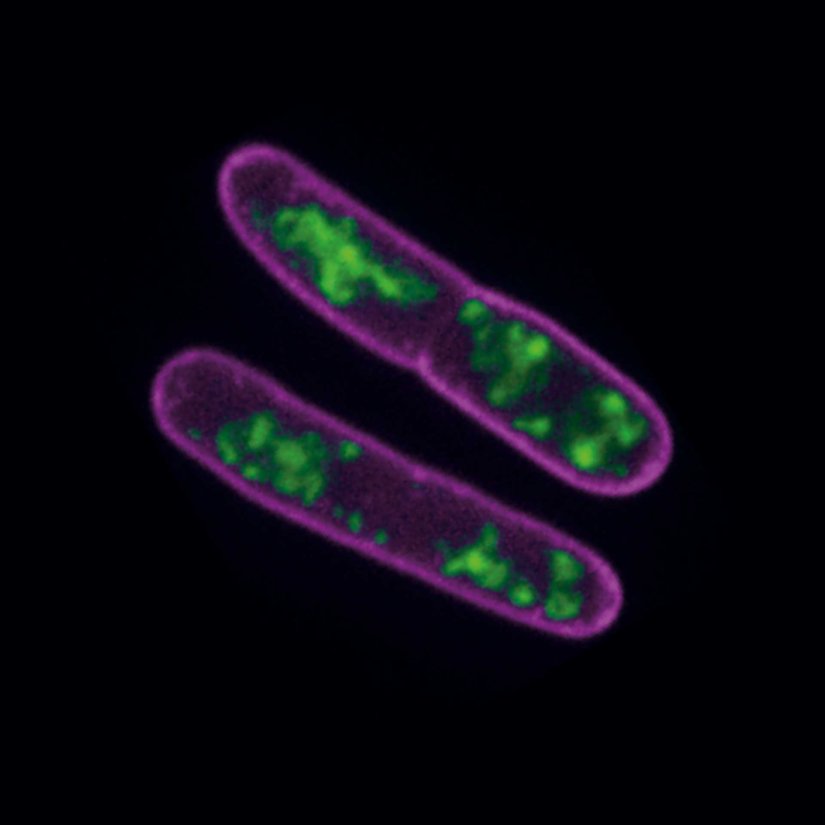
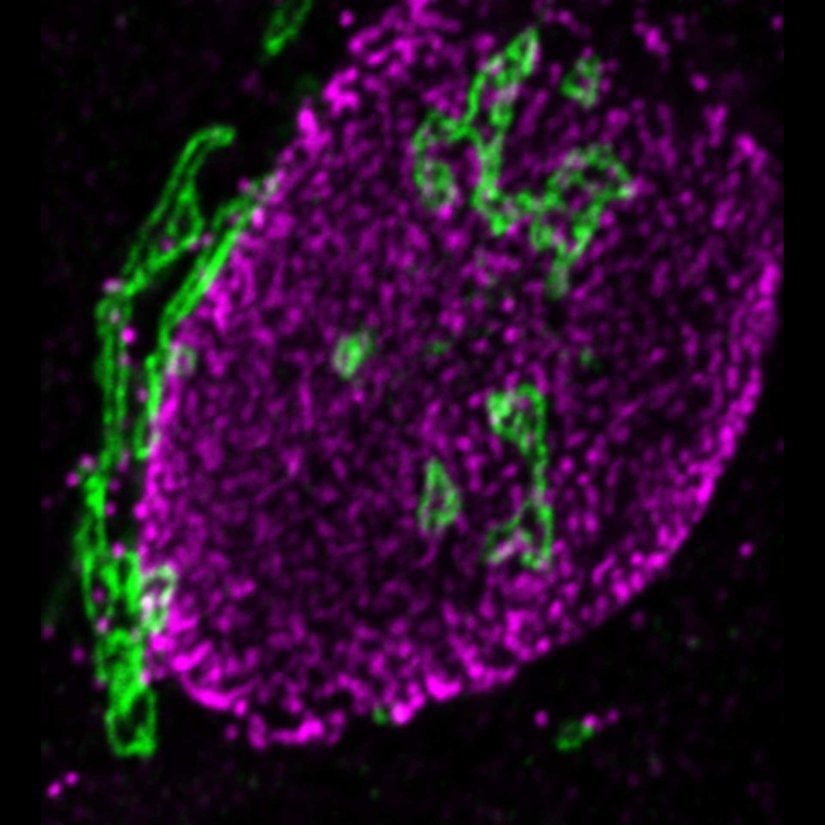
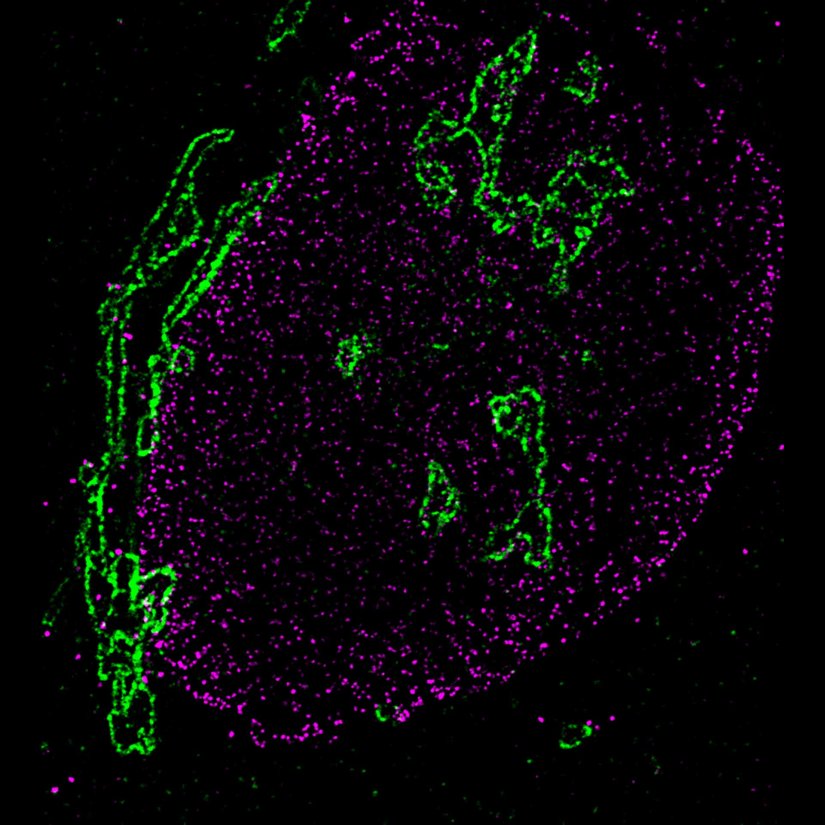
STED-PAINT imaging of bacterial membranes (magenta) and DNA (green) using a high concentration of the exchangeable labels.
Description
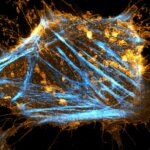
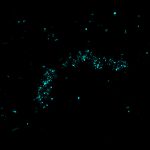
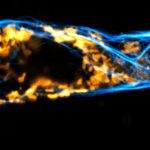
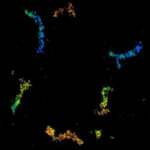
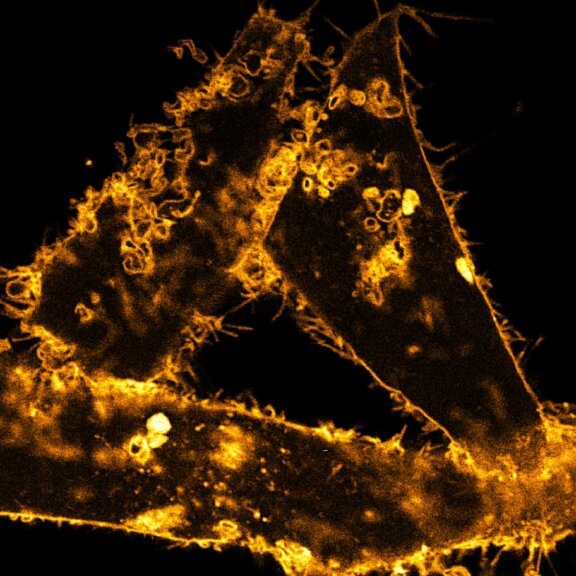
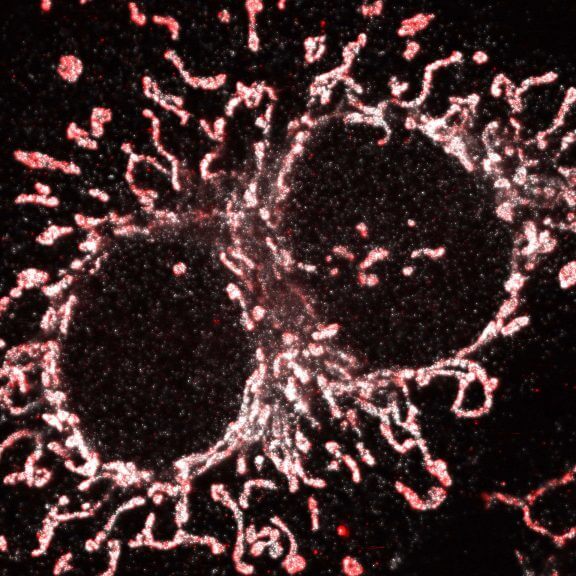
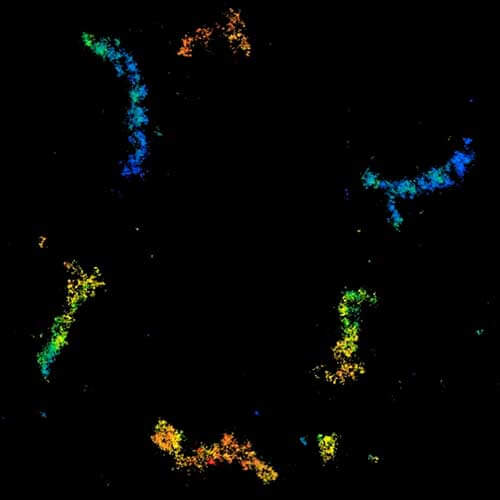
3D MINFLUX nanoscopy of Y. enterocolitica sorting platform protein Halo-YscL in xy view of two bacteria. The z-axis is color coded.
Sample from Alexander Carsten, Prof. Martin Aepfelbacher, Institute of Medical Microbiology, Virology and Hygiene, University Medical Center Hamburg Eppendorf, Germany
Description
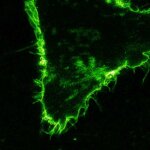
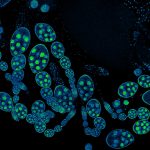
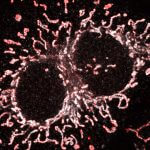
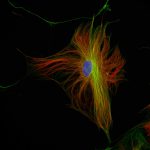
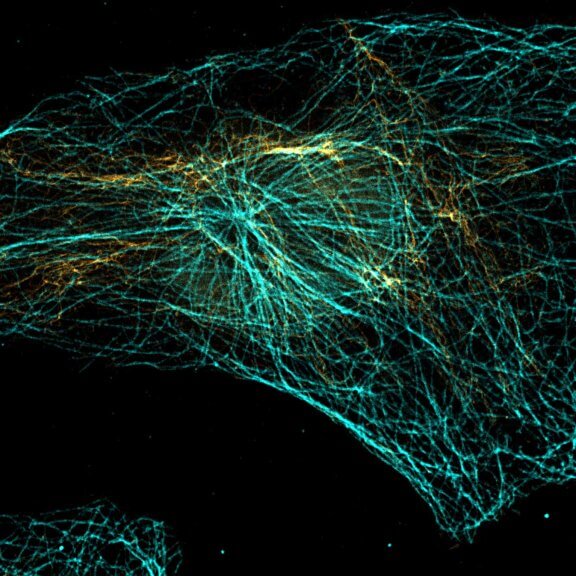
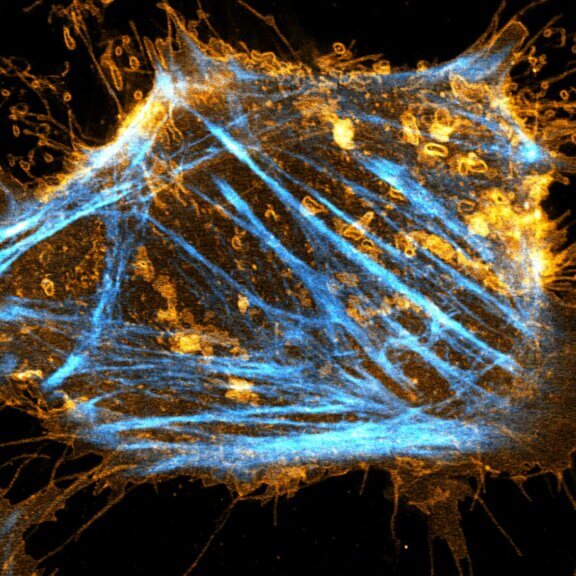
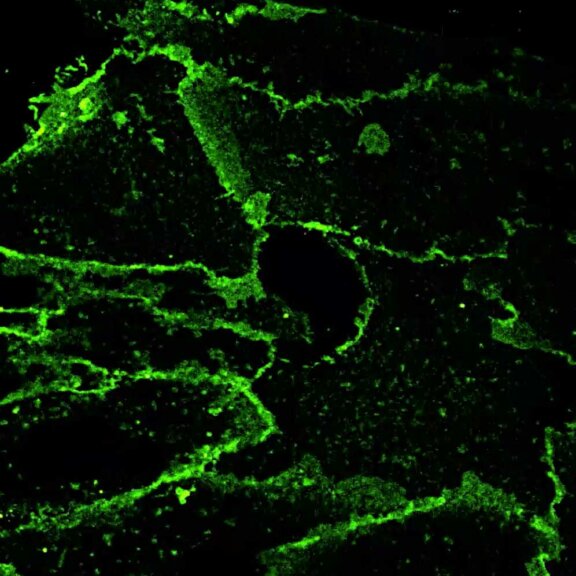
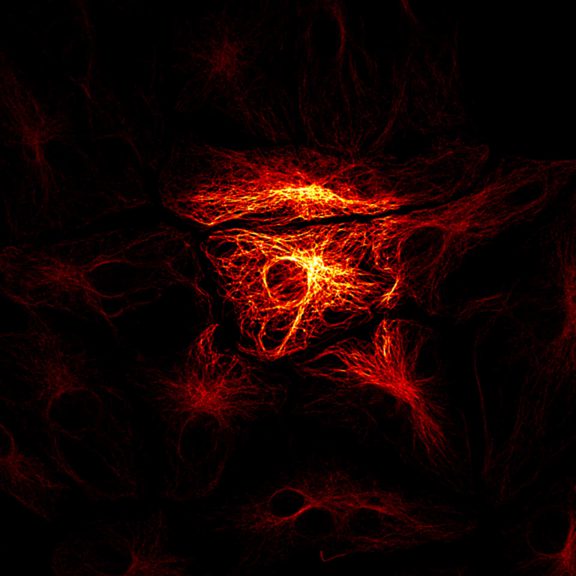
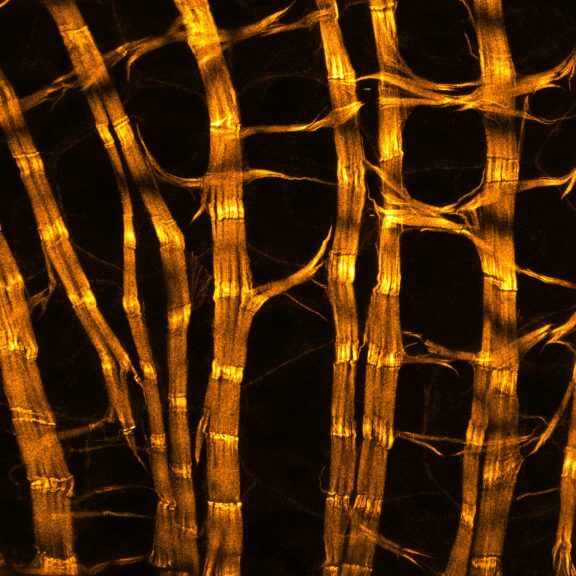
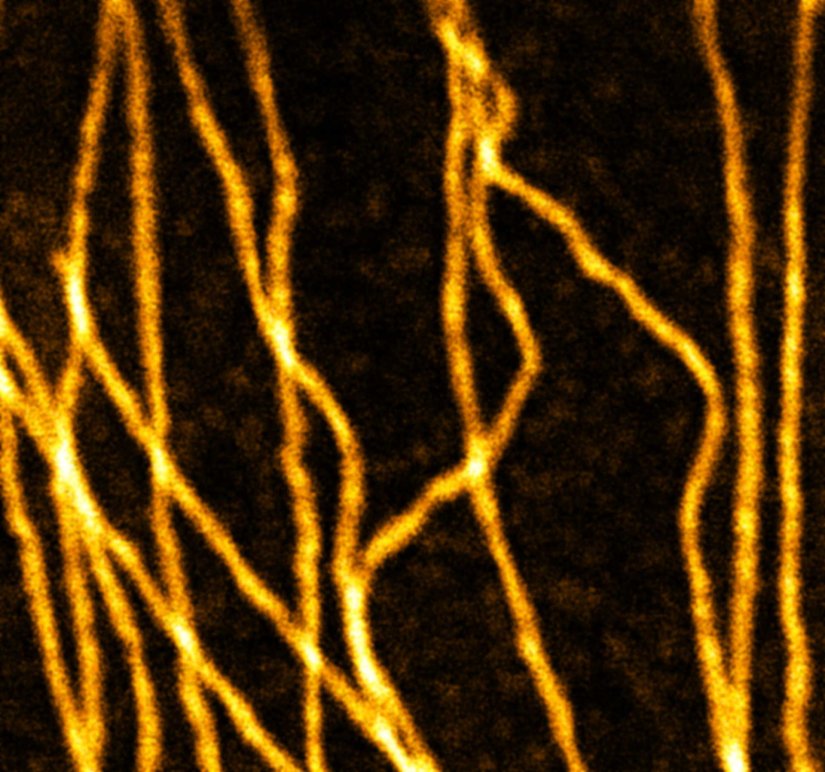
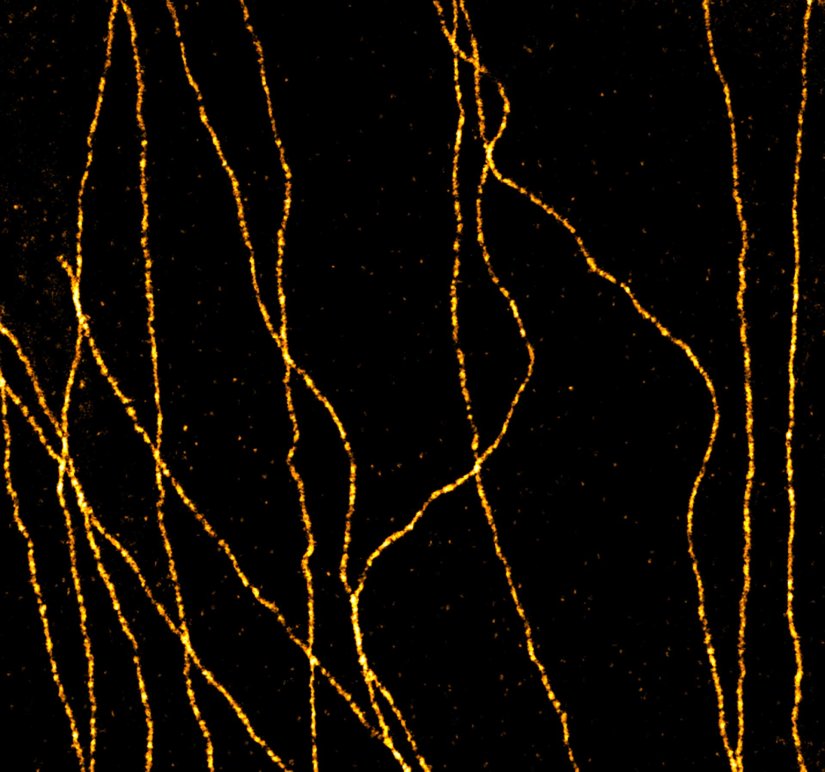
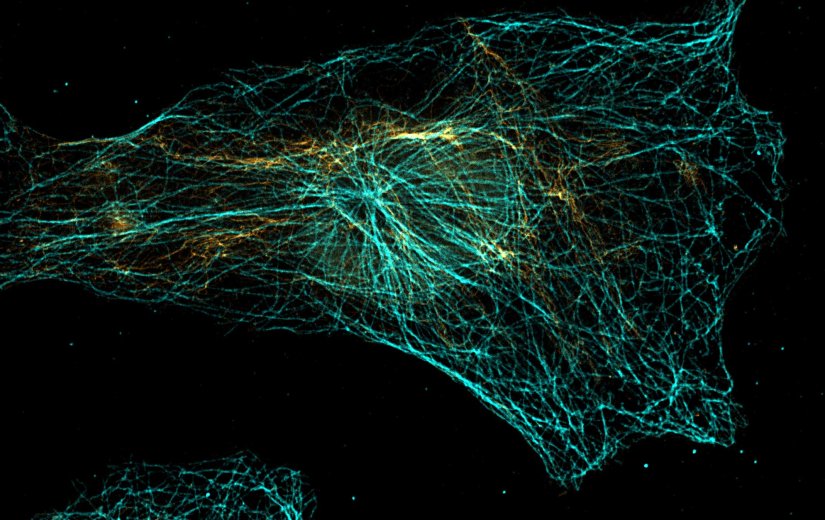
abberior STAR RED was coupled to polyclonal secondary nanobodies (alpaca VHH single domain antibodies) from Jackson ImmunoResearch and used to label tubulin in fixed mammalian cells via indirect immunofluorescence. The STED image was acquired with the STEDYCON system.
Description
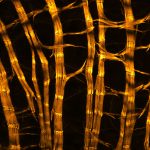
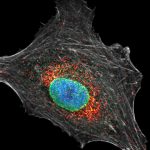
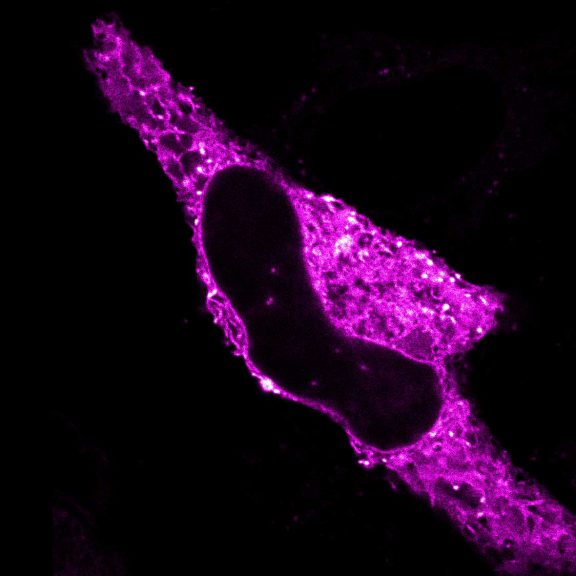
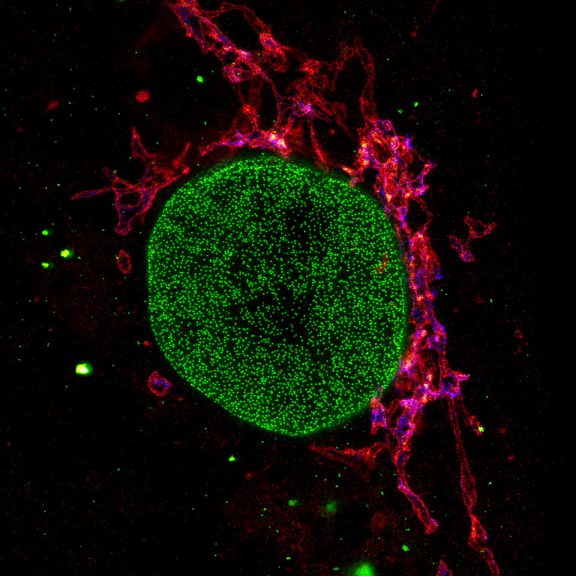
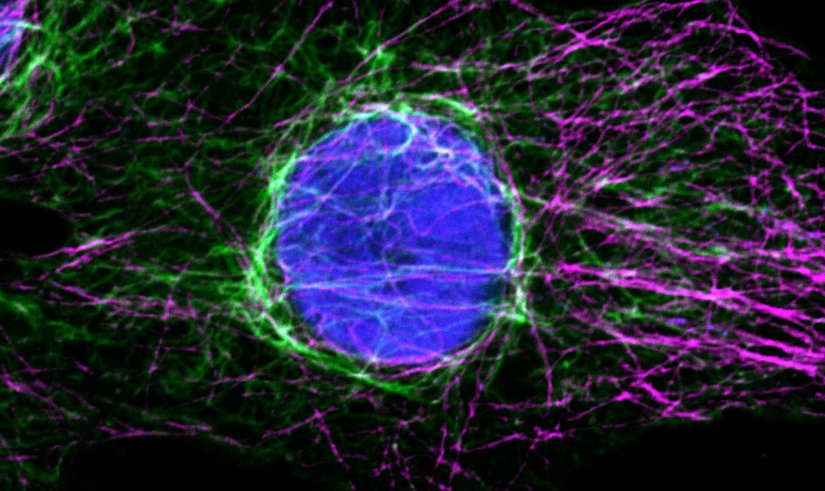
Proteins of the nuclear pore complex and the golgi apparatus were stained by indirect immunofluorescence using abberior STAR RED (nuclear pore complex, magenta) and abberior STAR 580 (golgi, green) coupled to polyclonal secondary nanobodies (alpaca VHH single domain antibodies) from Jackson ImmunoResearch. Images of the fixed mammalian cells were acquired with the STEDYCON.

Description
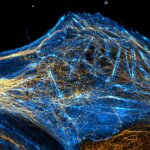
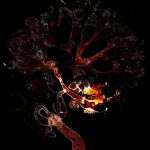
Vimentin and tubulin were stained by indirect immunofluorescence using abberior STAR RED (vimentin, orange) and abberior STAR 580 (tubulin, cyan) coupled to polyclonal secondary nanobodies (alpaca VHH single domain antibodies) from Jackson ImmunoResearch. Images of the fixed mammalian cells were acquired with the STEDYCON.
Description
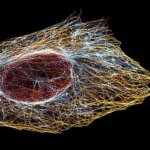
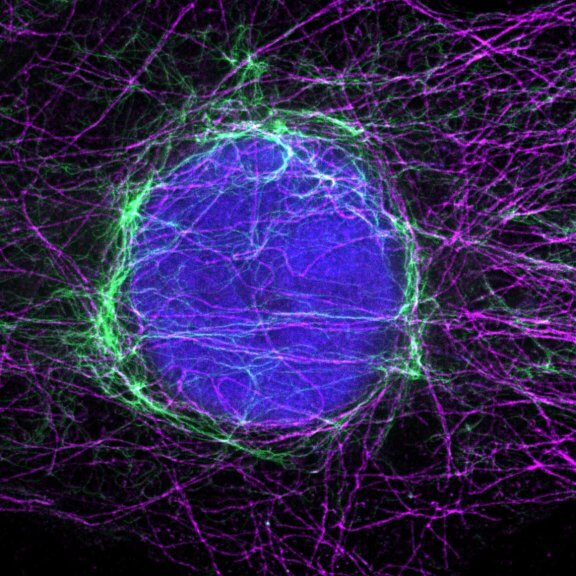
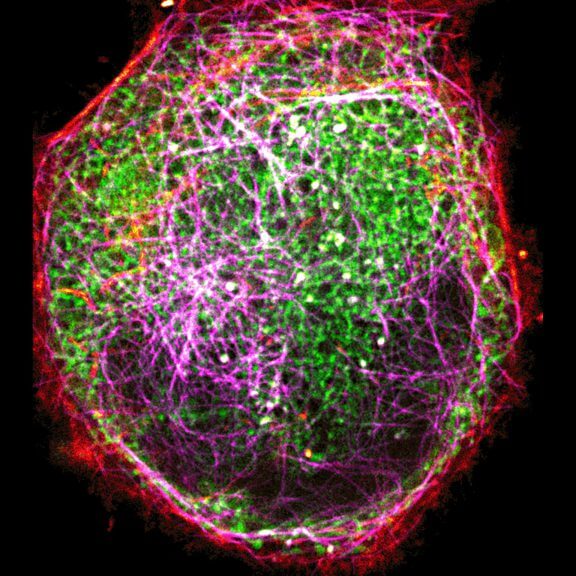
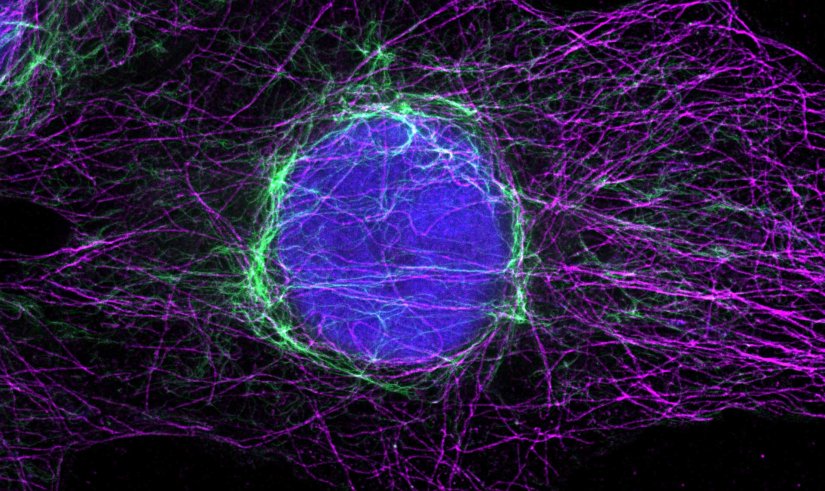
Indirect immunofluorescence staining with abberior STAR dyes coupled to polyclonal anti-mouse secondary nanobodies (alpaca VHH single domain antibodies) from Jackson ImmunoResearch shows excellent results in 3-color STED imaging. Staining was performed according to the premix & stain protocol. Vimentin (abberior STAR RED, green), tubulin (abberior STAR 580, magenta) and dsDNA (abberior STAR 460L, blue) were labeled in fixed mammalian cells and visualized with the STEDYCON.
Description
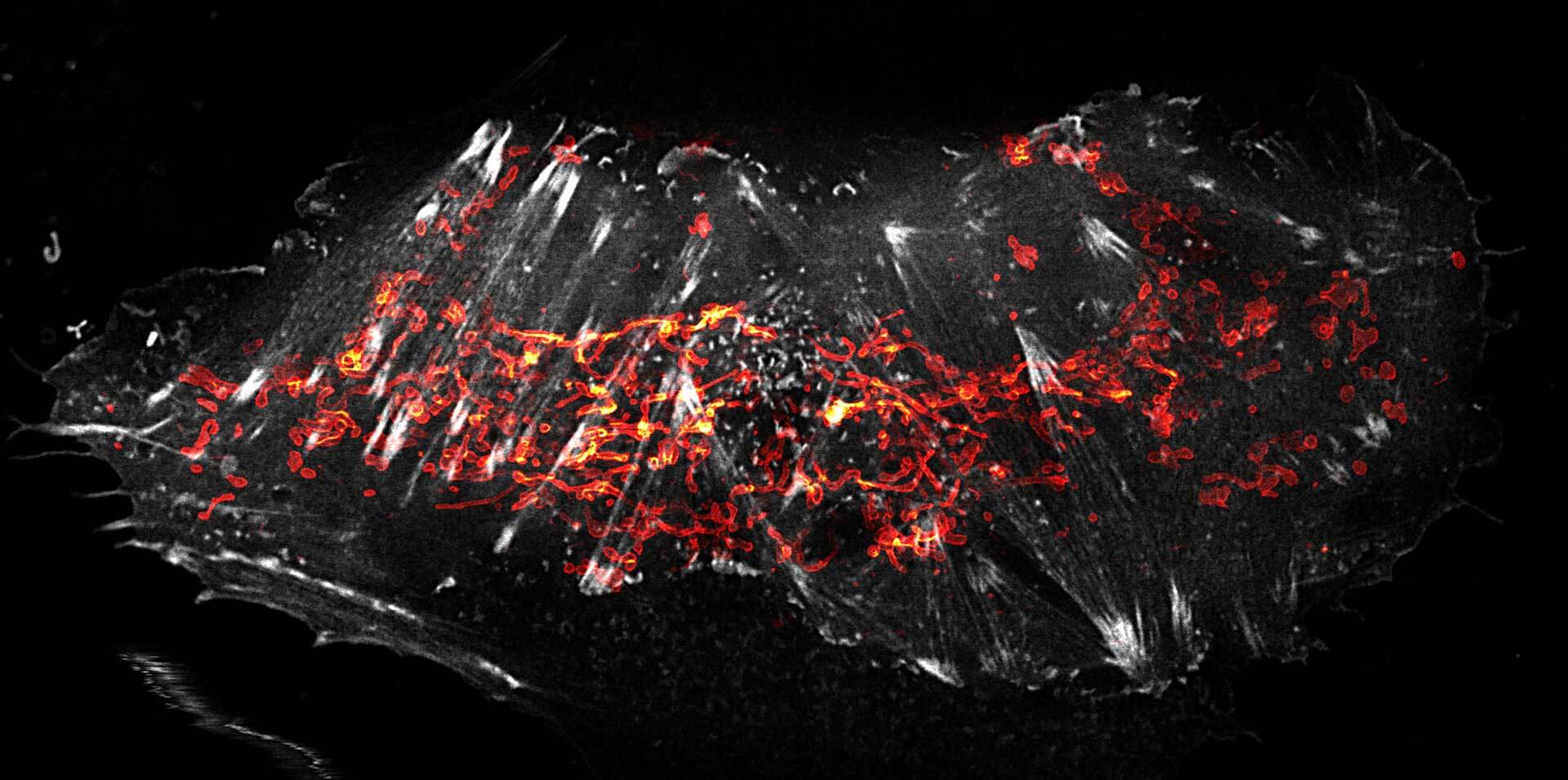
OMP25-SNAP protein in mitochondria labeled with abberior LIVE 610 SNAP (magenta) and Actin^K118TAG labeled with abberior LIVE 550 click (green). Movie with 60 frames.
Description
Description
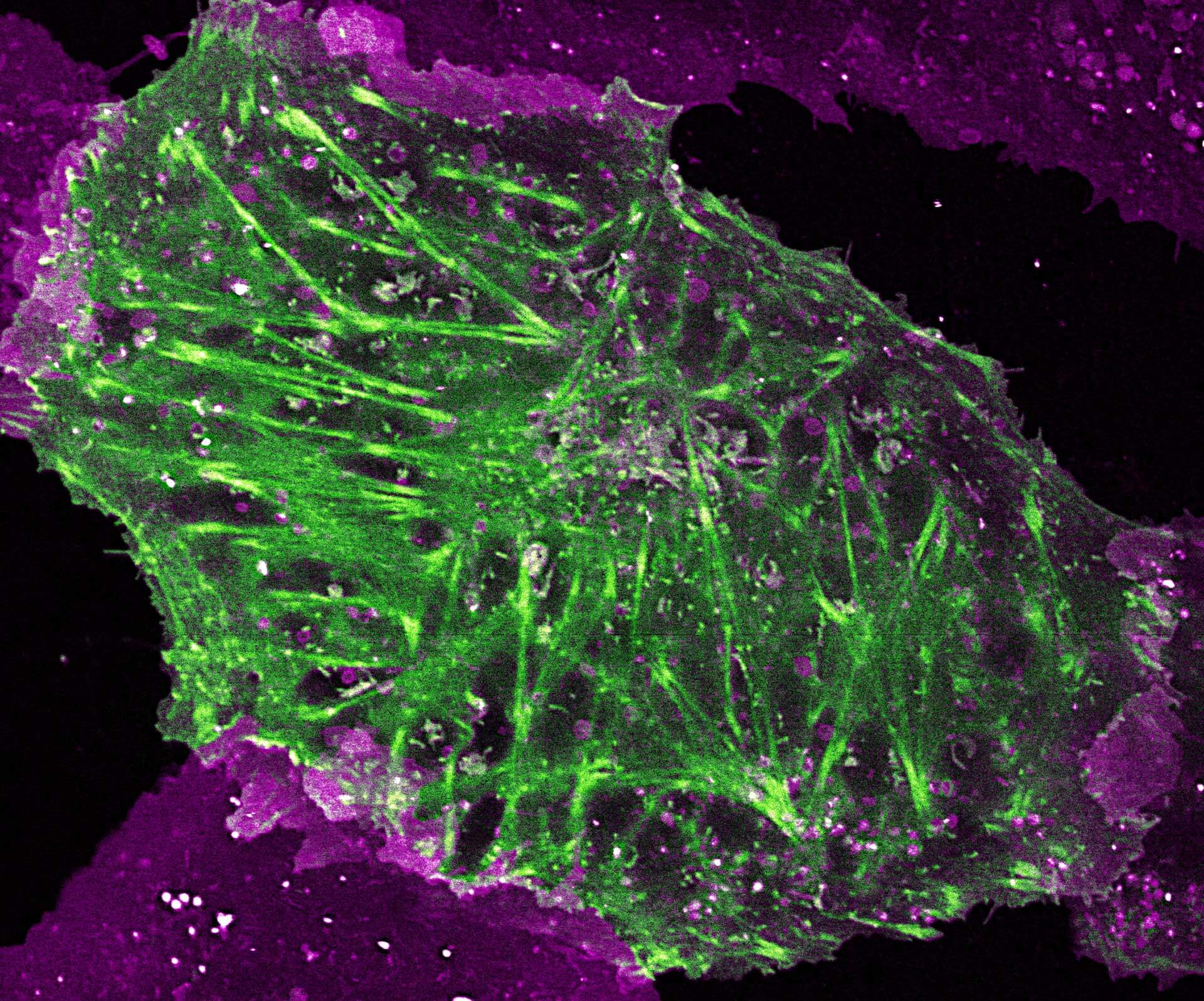
2-color STED image of a living cell expressing actin K118TAG incorporating TCO*A labeled with abberior LIVE 590 click (green). Plasma membrane is highlighted with abberior STAR RED membrane (magenta).
Description
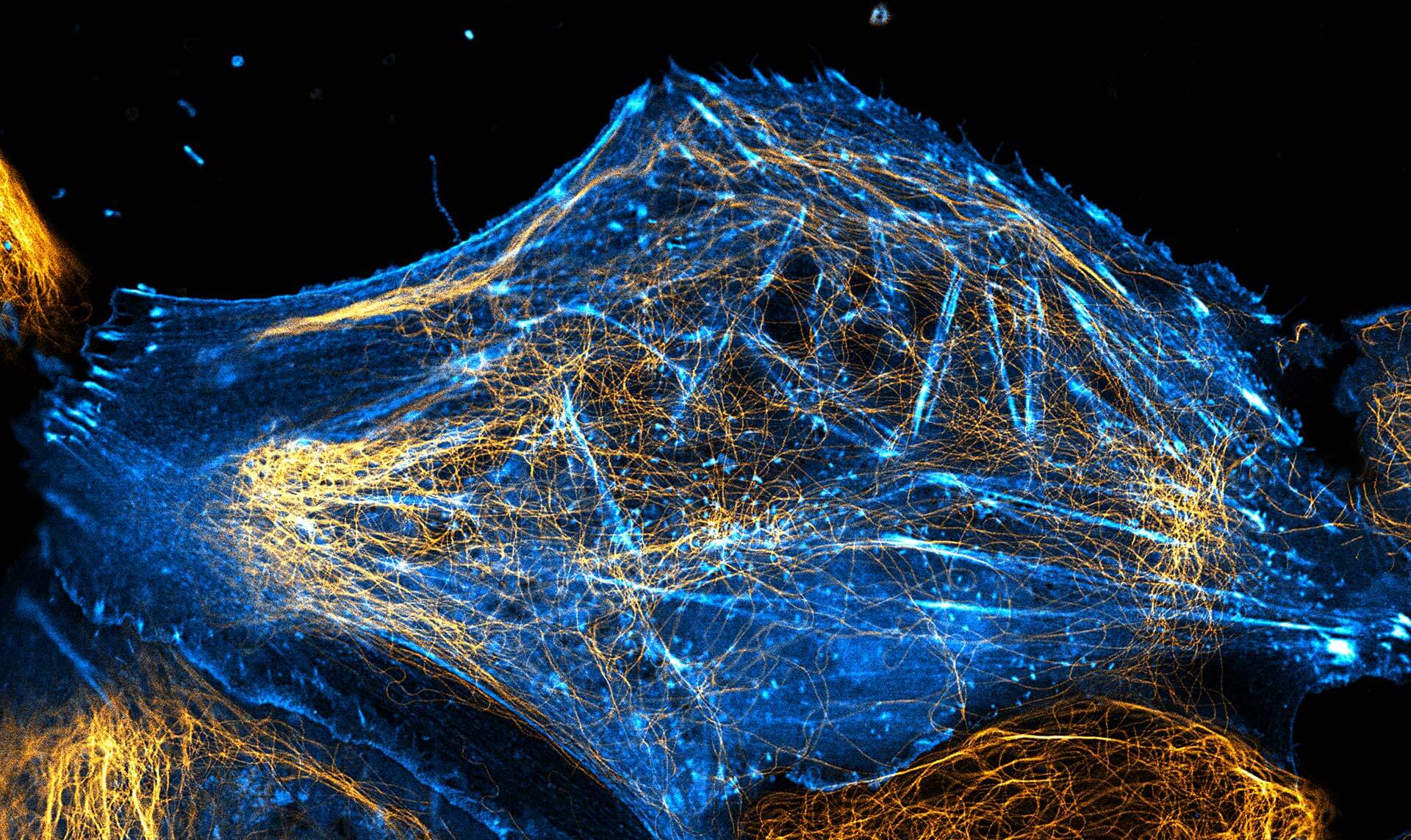
2-color STED image of a living cell expressing actinK118TAG incorporating TCO*A labeled with abberior LIVE 550 click (cyan). Tubulin filaments were stained with our direct probe abberior LIVE 610 tubulin (yellow).
Description
Perfect 3-color combination for STED @ 775 nm. abberior STAR RED NUP98 (red) and STAR ORANGE vimentin (cyan) complemented by the new STAR 460L tubulin (yellow). Cultured mammalian cells were fixed and labeled by indirect immunofluorescence.
Description
We are happy to introduce the first commercial long Stokes-shift dye for live-cell imaging applications. The NEW abberior LIVE 460L makes 3-color live-cell STED at 775 nm possible!
Live-cell STED image of a cell expressing a SNAP-tag® in an outer mitochondrial membrane protein visualized by our abberior LIVE 460L SNAP (orange). Tubulin filaments are highlighted with LIVE 610 (cyan) and the DNA with LIVE 560 DNA (red).
Description
STED time-lapse of living mammalian cell stained with abberior STAR ORANGE membrane.
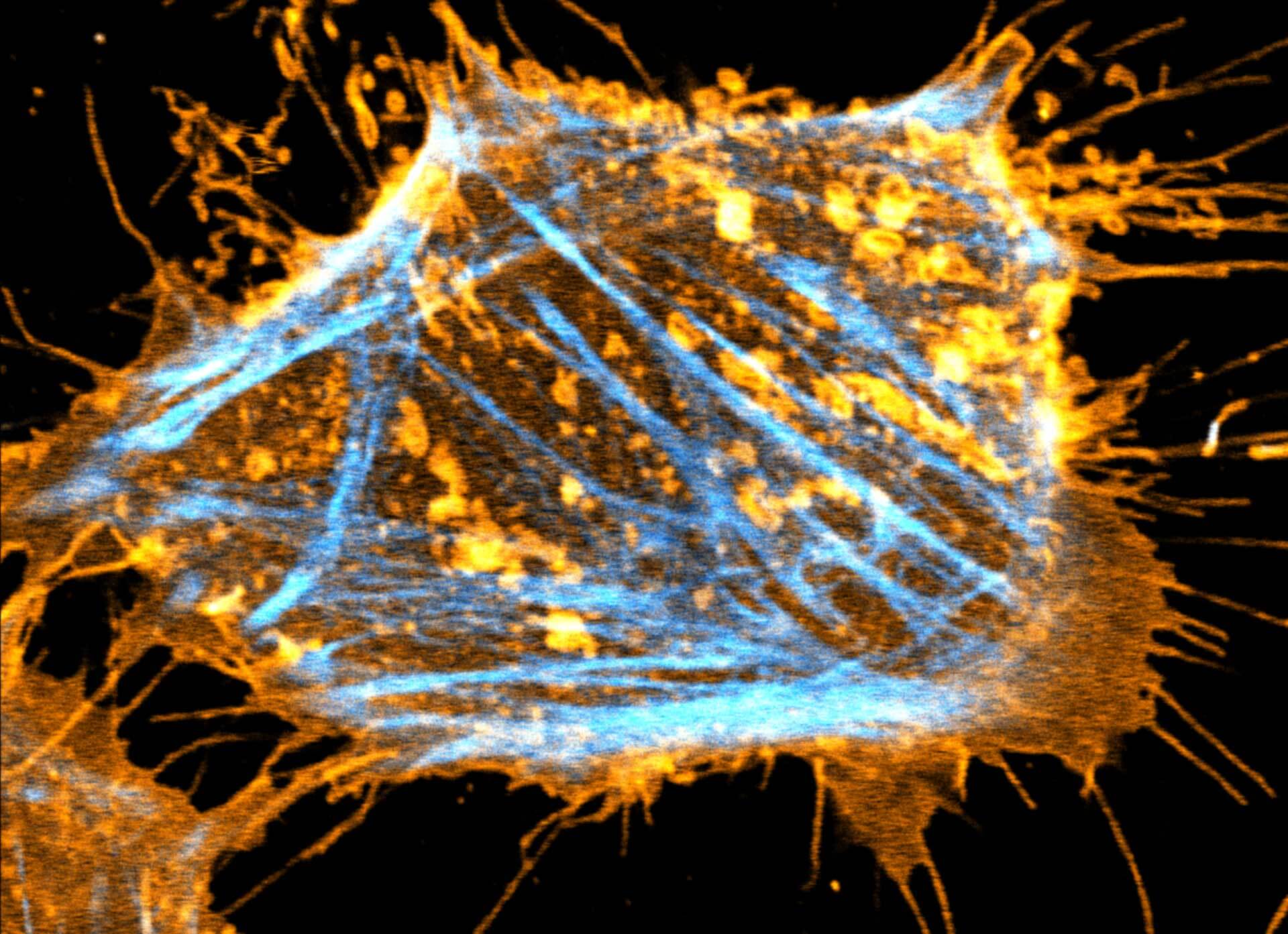
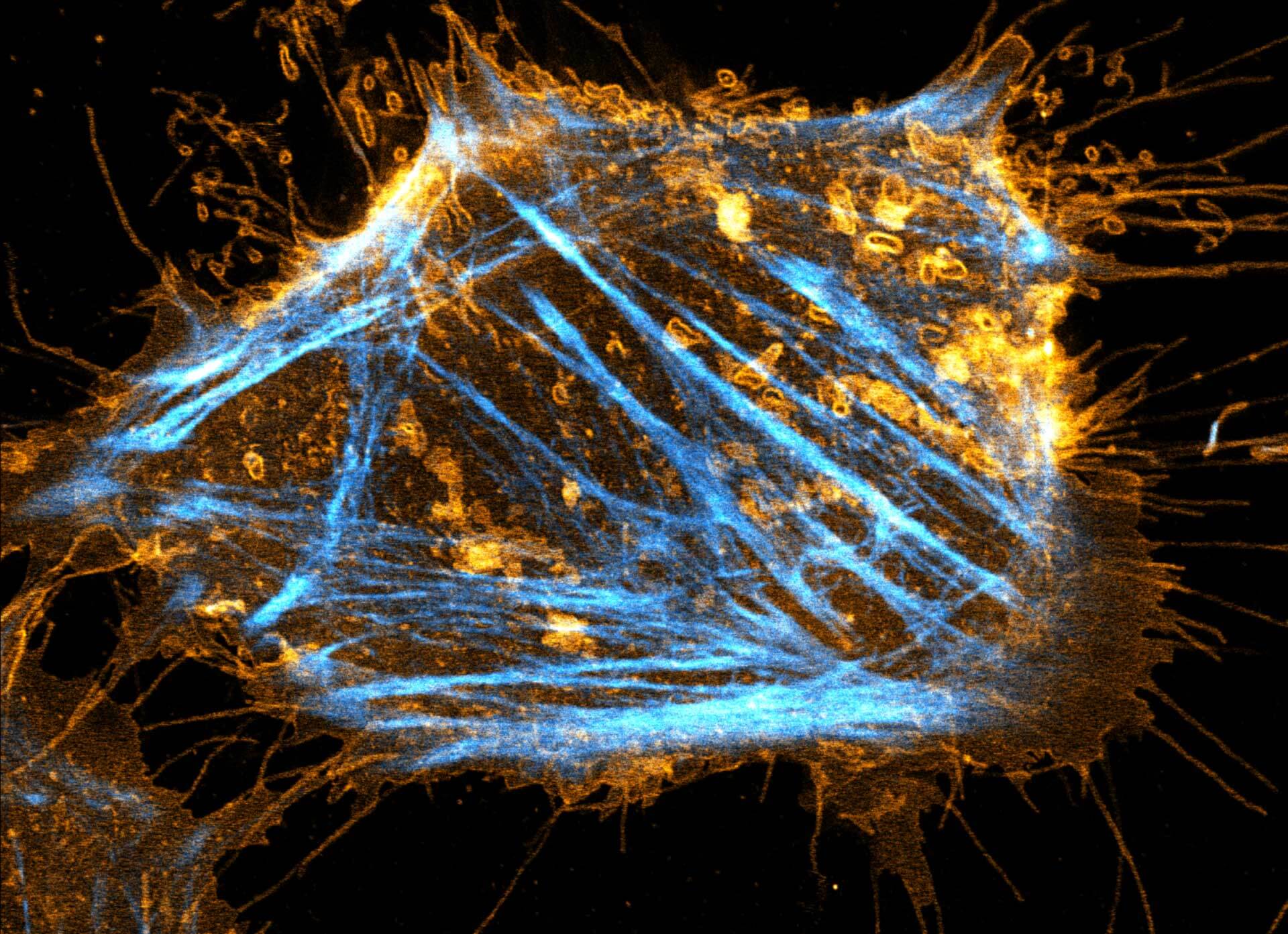
Description
Two color live-cell STED and confocal image of a mammalian cultured cell stained with abberior STAR RED membrane (orange) and abberior LIVE 590 actin (cyan).
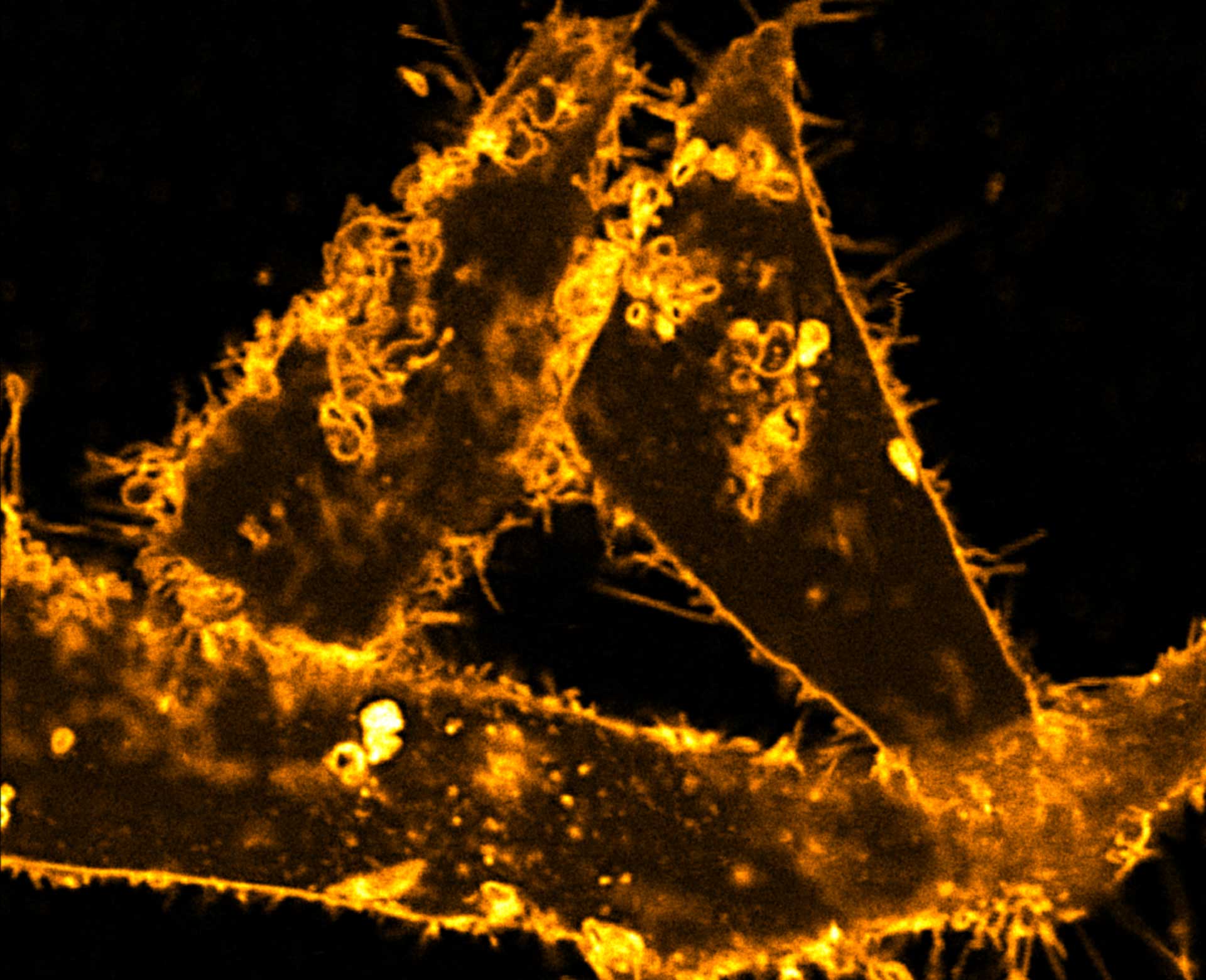
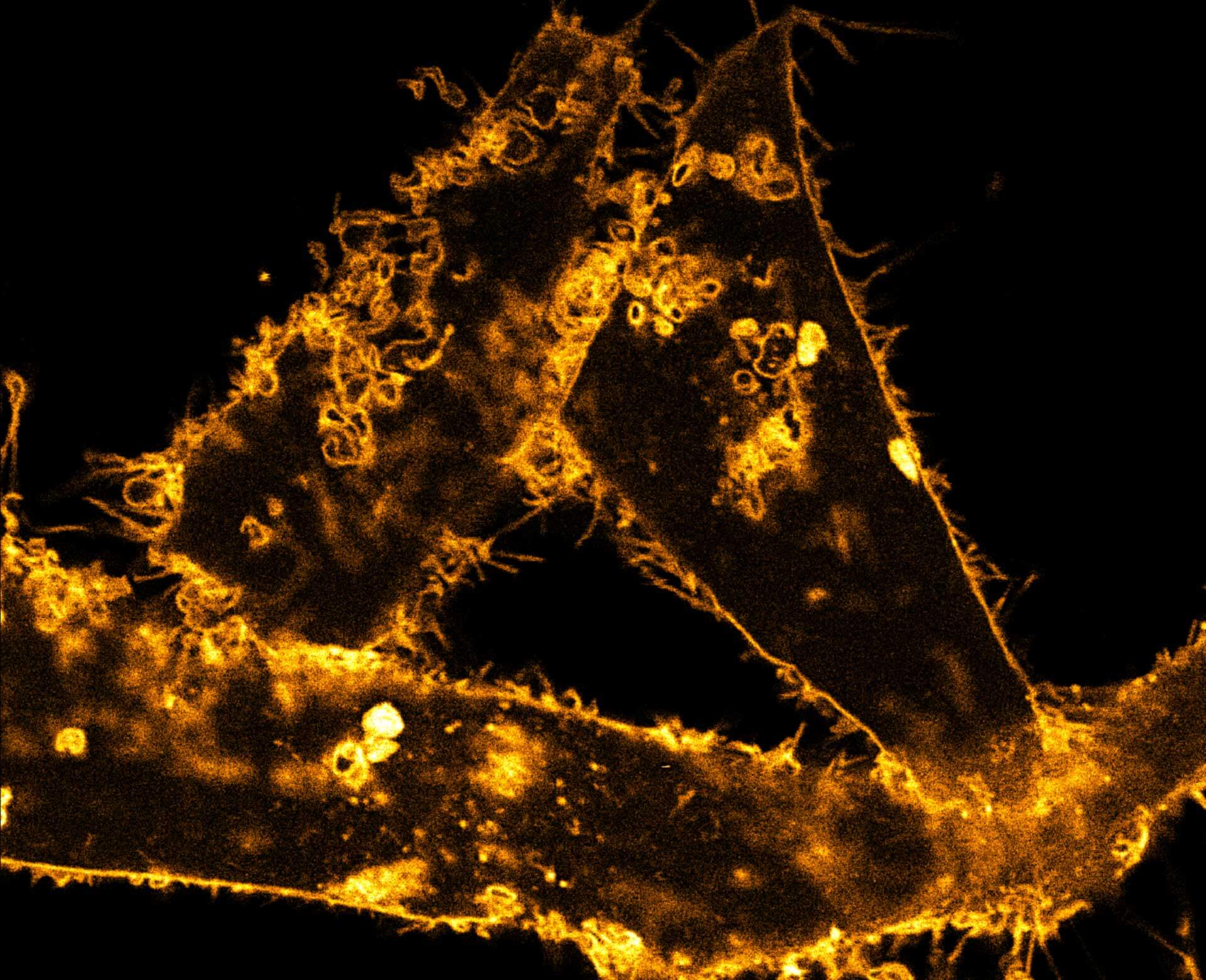
Description
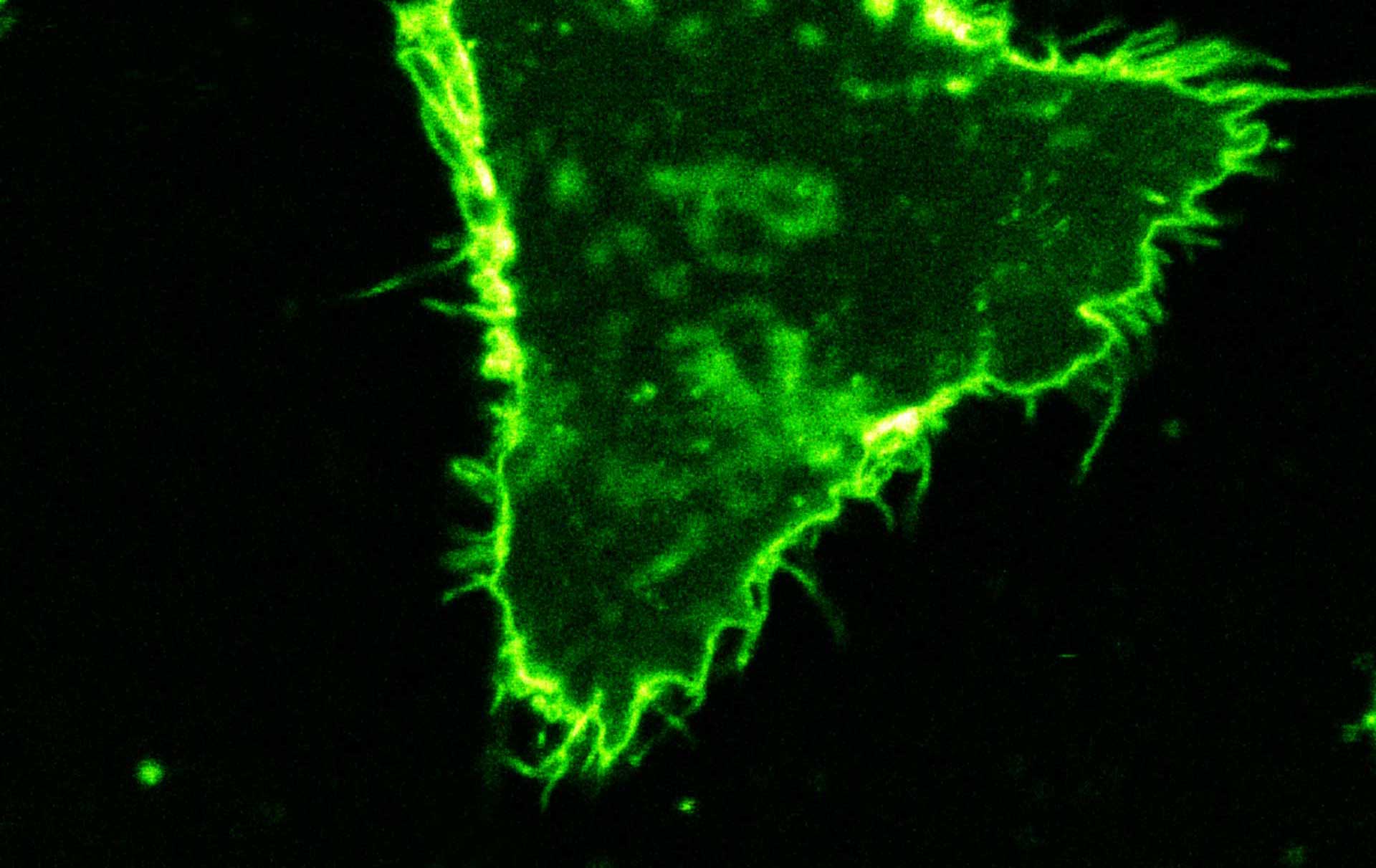
Comparison of STED and confocal of a living mammalian cell stained with abberior STAR ORANGE membrane.
Description
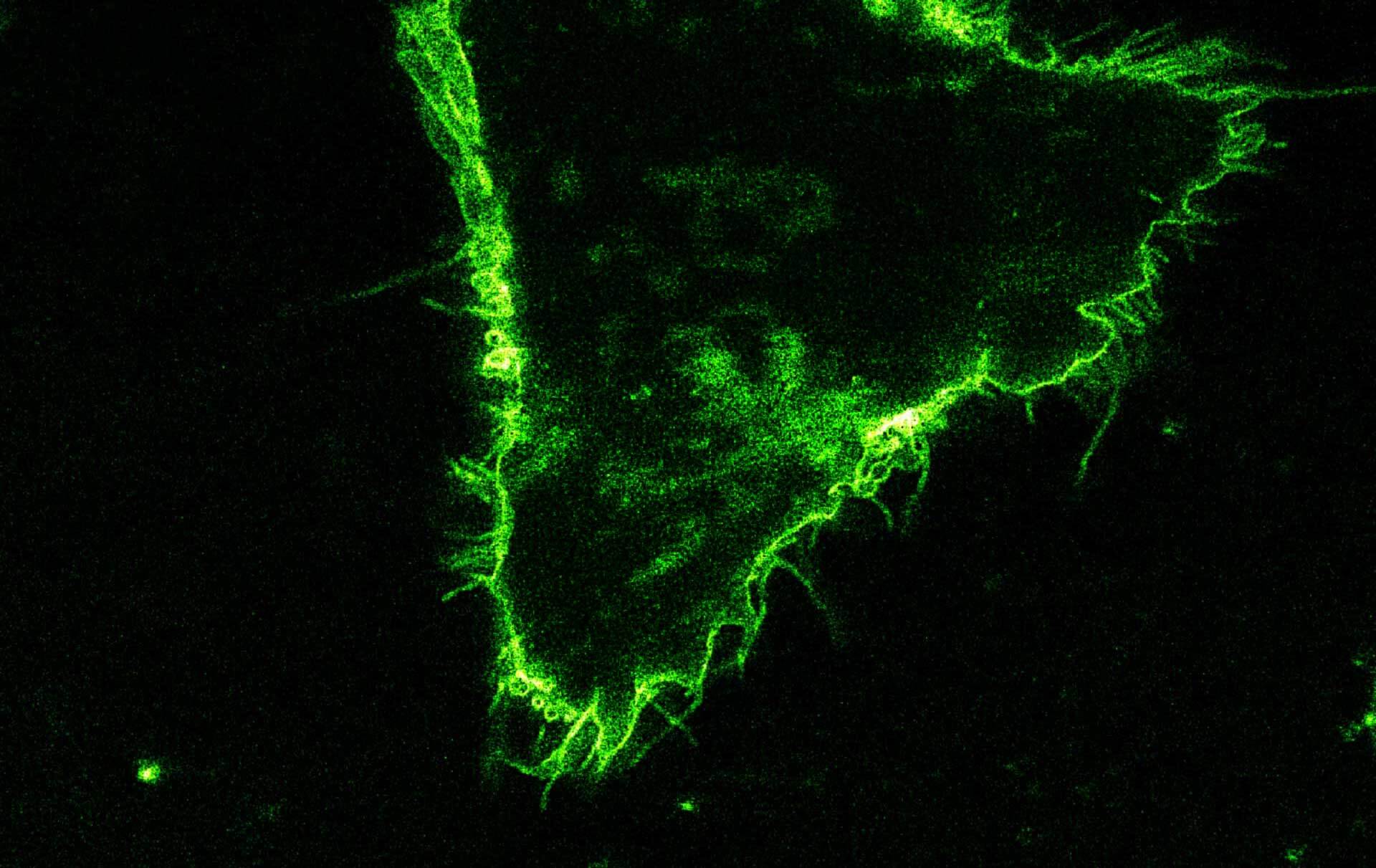
Comparison of STED and confocal of a living mammalian cell stained with abberior STAR 488 membrane. Images were acquired with our FACILITY and STED @ 595 nm.
Description
Confocal time-lapse of living mammalian cell stained with abberior STAR 488 membrane.
Description
abberior CAGE 552 is a caged dye which is initially nonfluorescent and colorless substance which, upon photolysis with UV light, readily transforms irreversibly into a red and highly fluorescent dye.
The cytoskeleton is visible after UV induced uncaging of abberior CAGE 552.
Description
3D MINFLUX image of the mitochondrial import receptor Tom20 labeled with abberior FLUX 647 in fixed mammalian cells using indirect immunolabeling. Please note that Tom20 is only localized at the mitochondrial surface. The axial coordinate is color-coded.
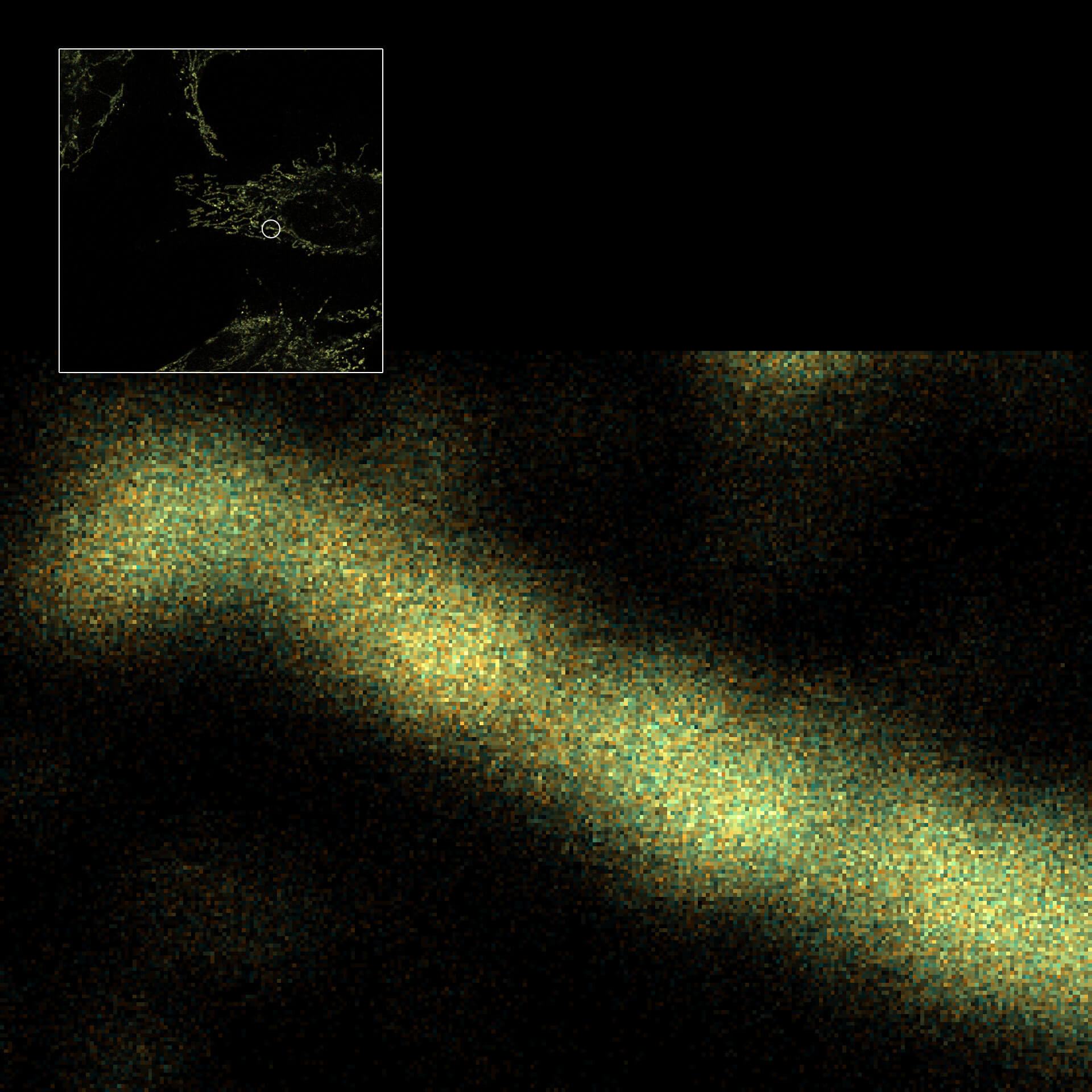
Description
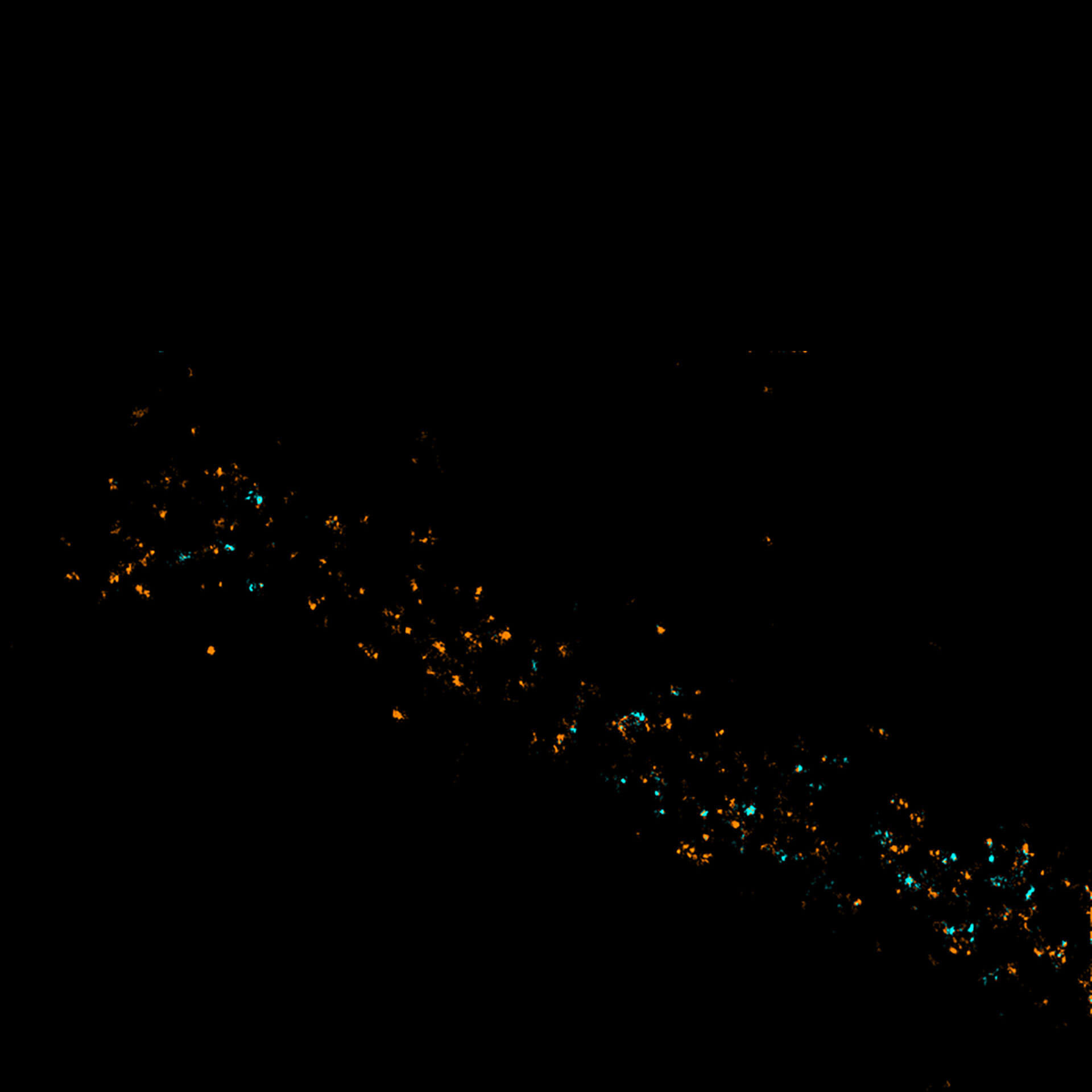
Two-color MINFLUX revealing an inner and outer mitochondrial membrane marker. Cultured mammalian cell labeled with indirect immunofluorescence using secondary antibodies coupled to abberior FLUX 640 (orange) and FLUX 680 (cyan). MINFLUX enables the visualization and separation of both structures.

Description
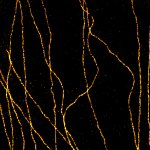
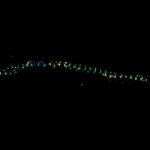
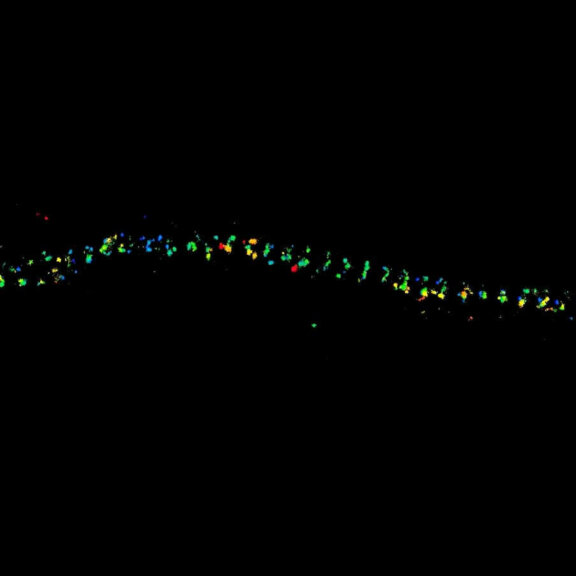
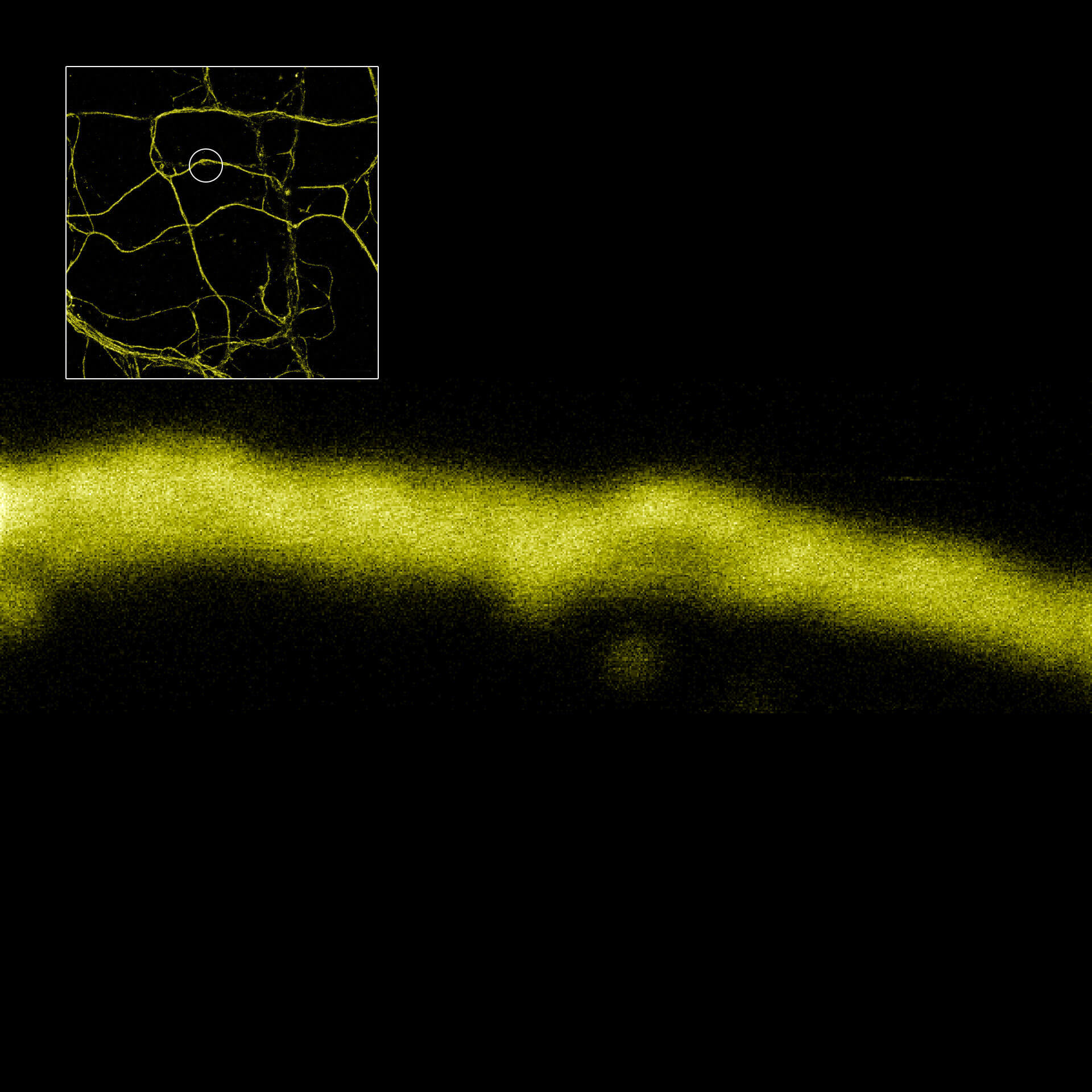
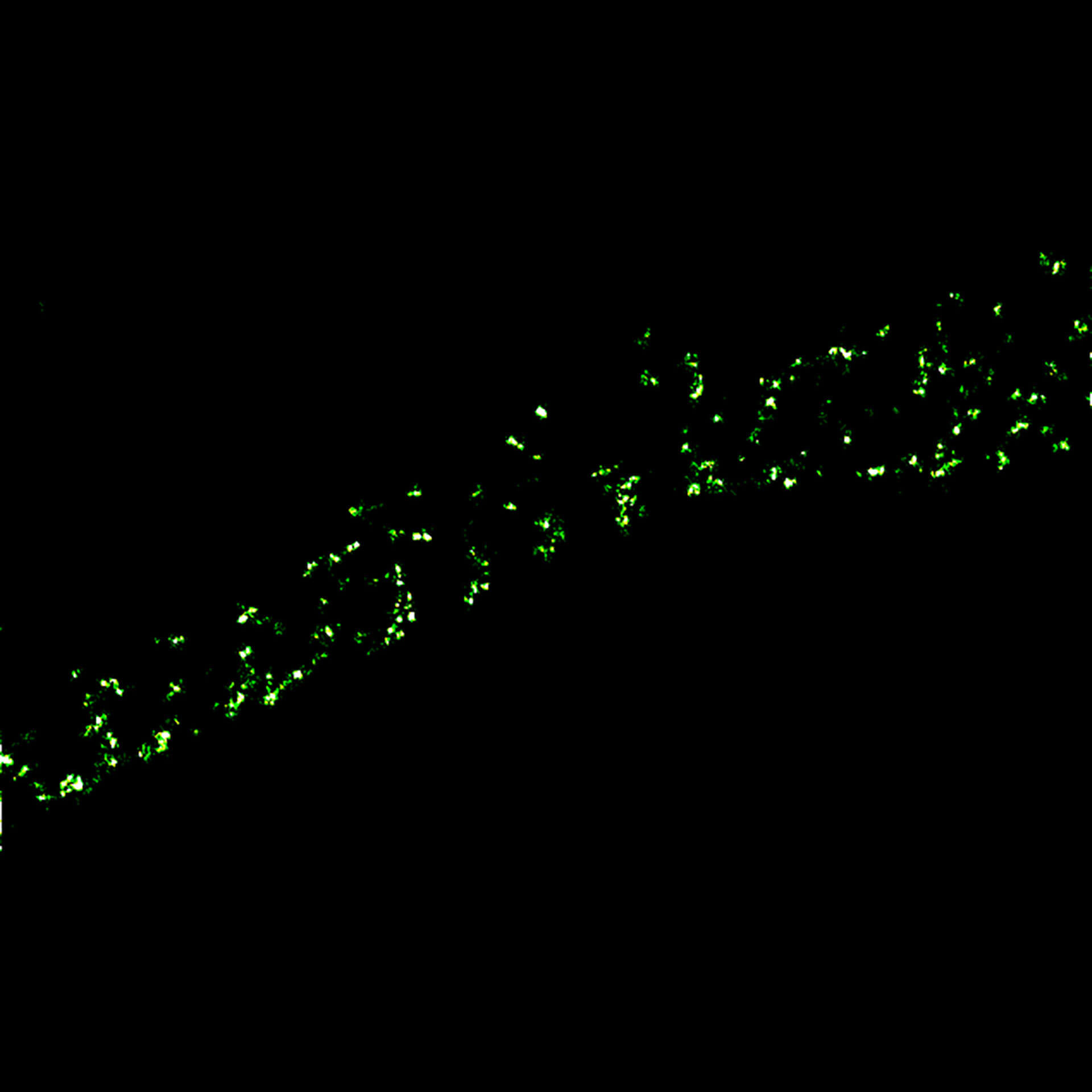
MINFLUX imaging of βII spectrin in a primary hippocampal neuron labeled with abberior FLUX 680 by indirect immunofluorescence. Please note the periodic arrangement of spectrin along the axon.
Description
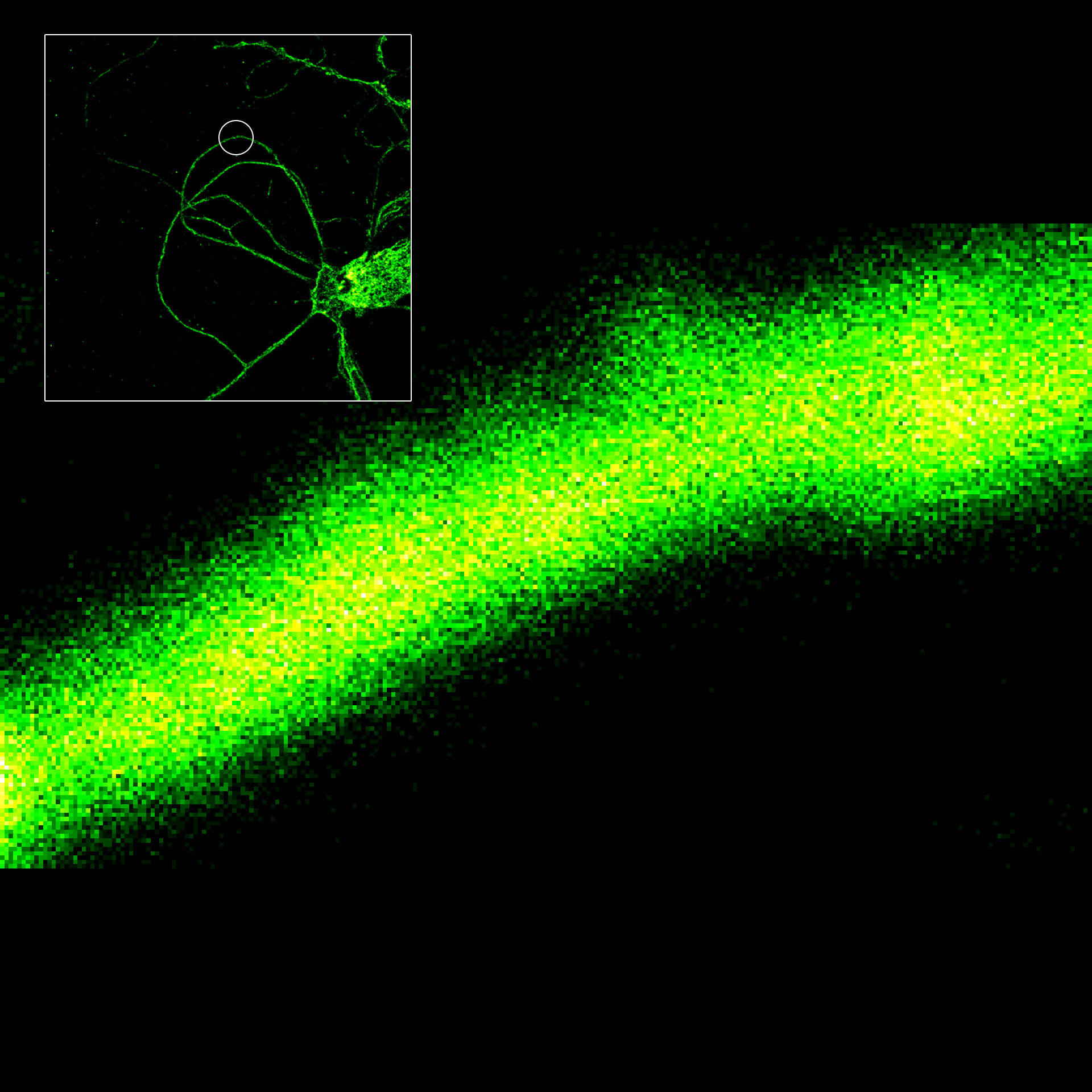
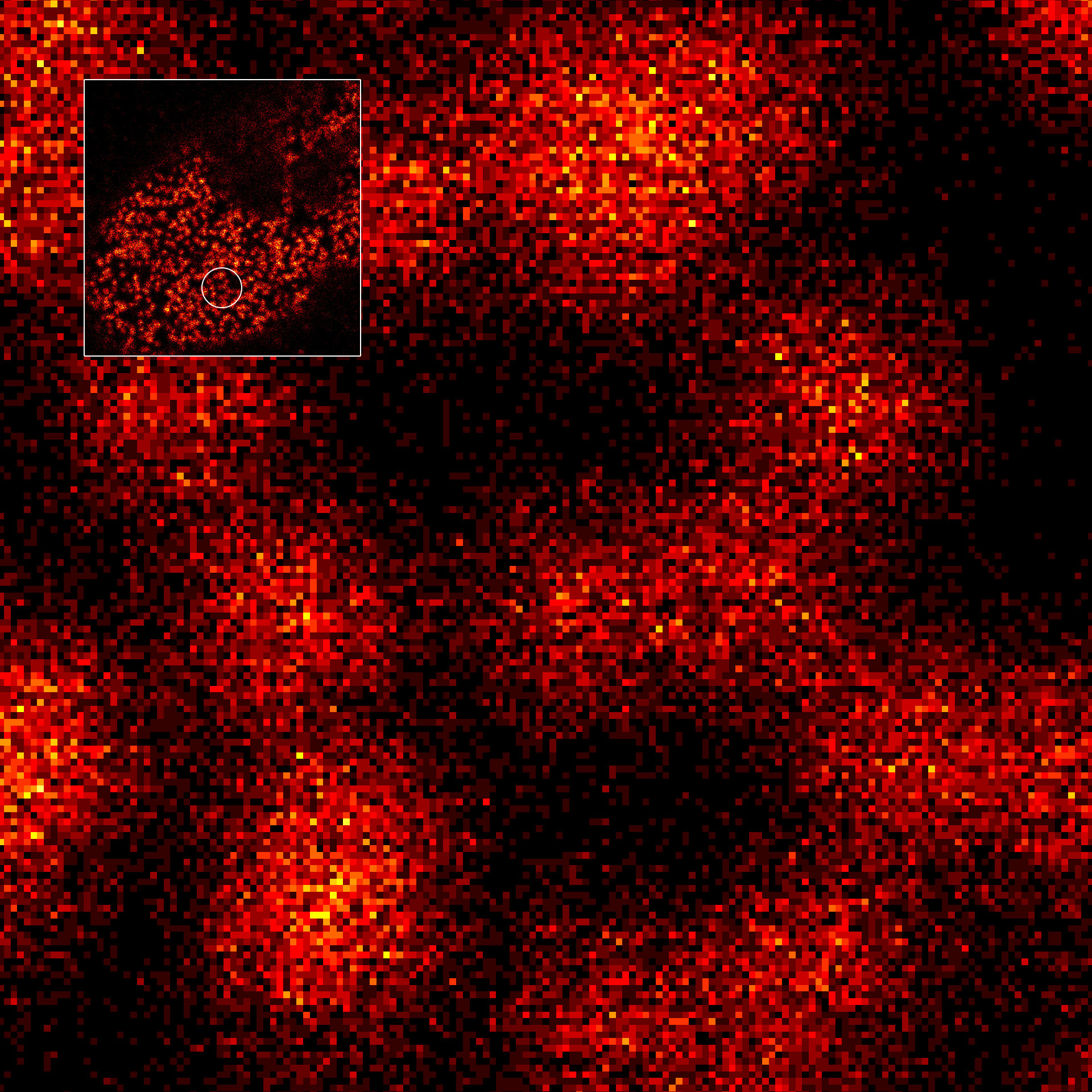
MINFLUX image of axonal βII spectrin labeled with abberior FLUX 660 in primary hippocampal neurons. Note the periodic arrangement of spectrin along the axon, and the absence of any details in the confocal counterpart image.

Description
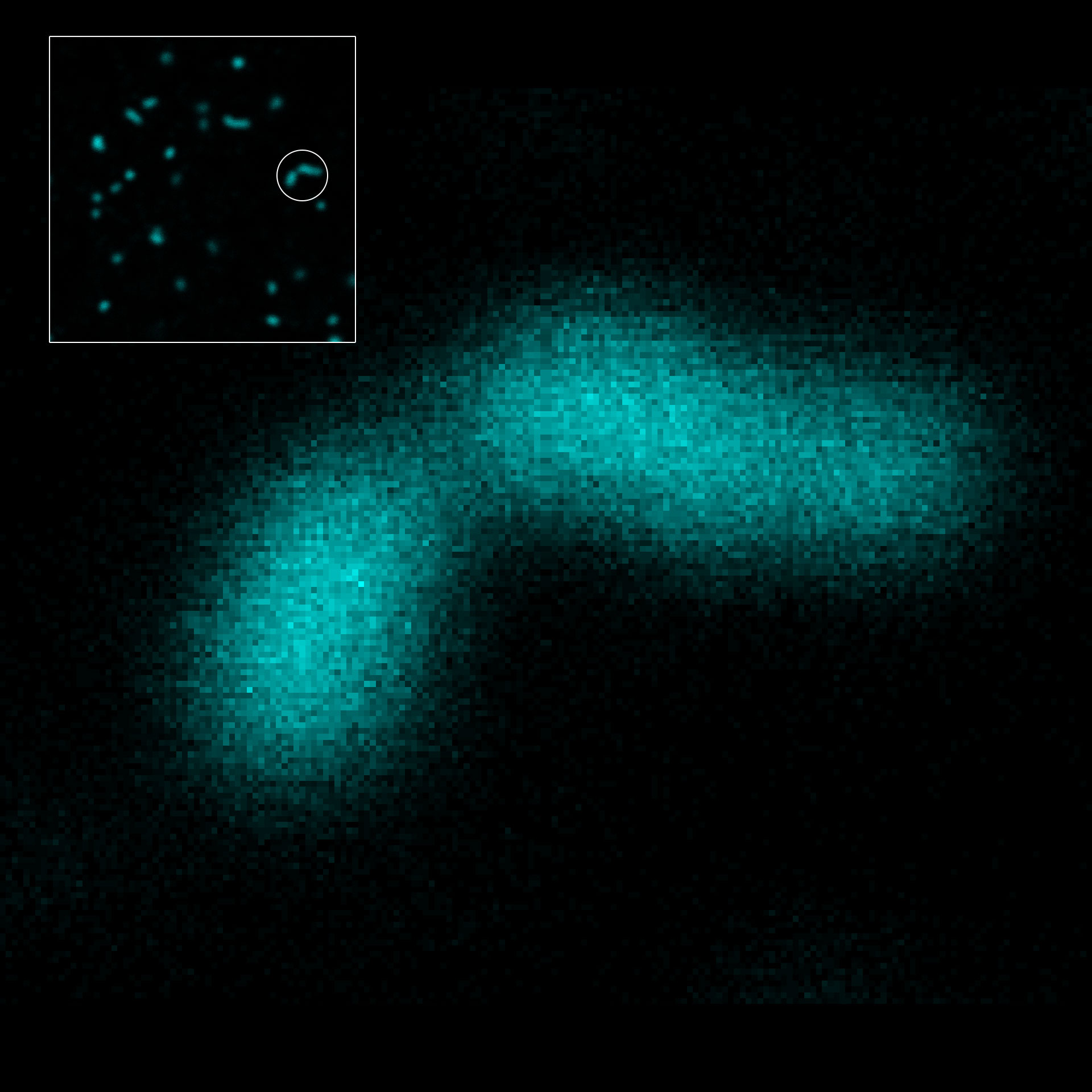
2D MINFLUX nanoscopy of the nuclear pore complex subunits. Fixed mammalian cells expressing SNAP-tag® NUP96 were labeled with abberior FLUX 647 SNAP. In contrast to confocal microscopy, 2D MINFLUX allows to visualize the shape and arrangement of individual subunits of the nuclear pore complex.
Description
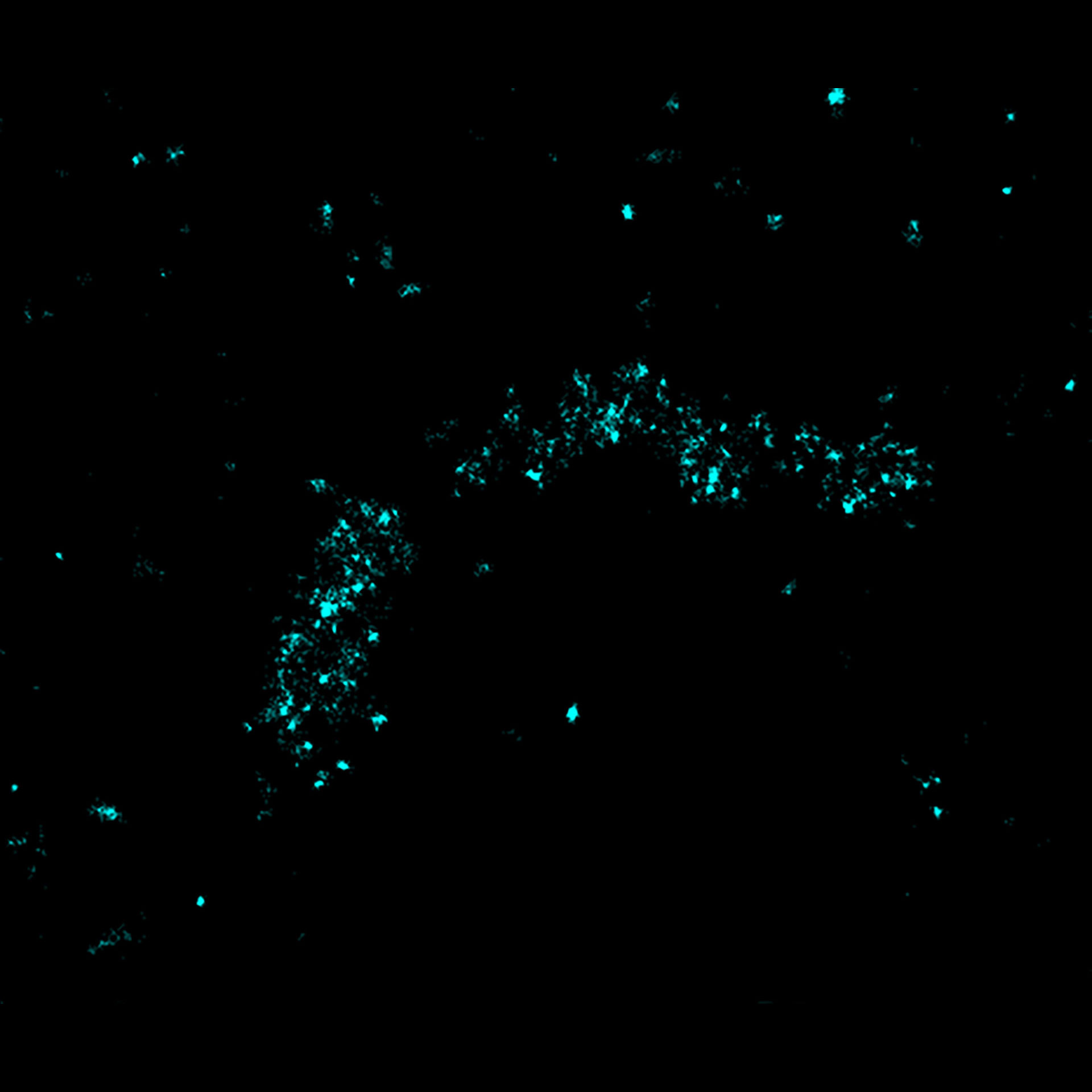
2D MINFLUX imaging of the peroxisomal membrane protein PMP70 labeled with abberior FLUX 640 in fixed mammalian cells using indirect immunofluorescence.
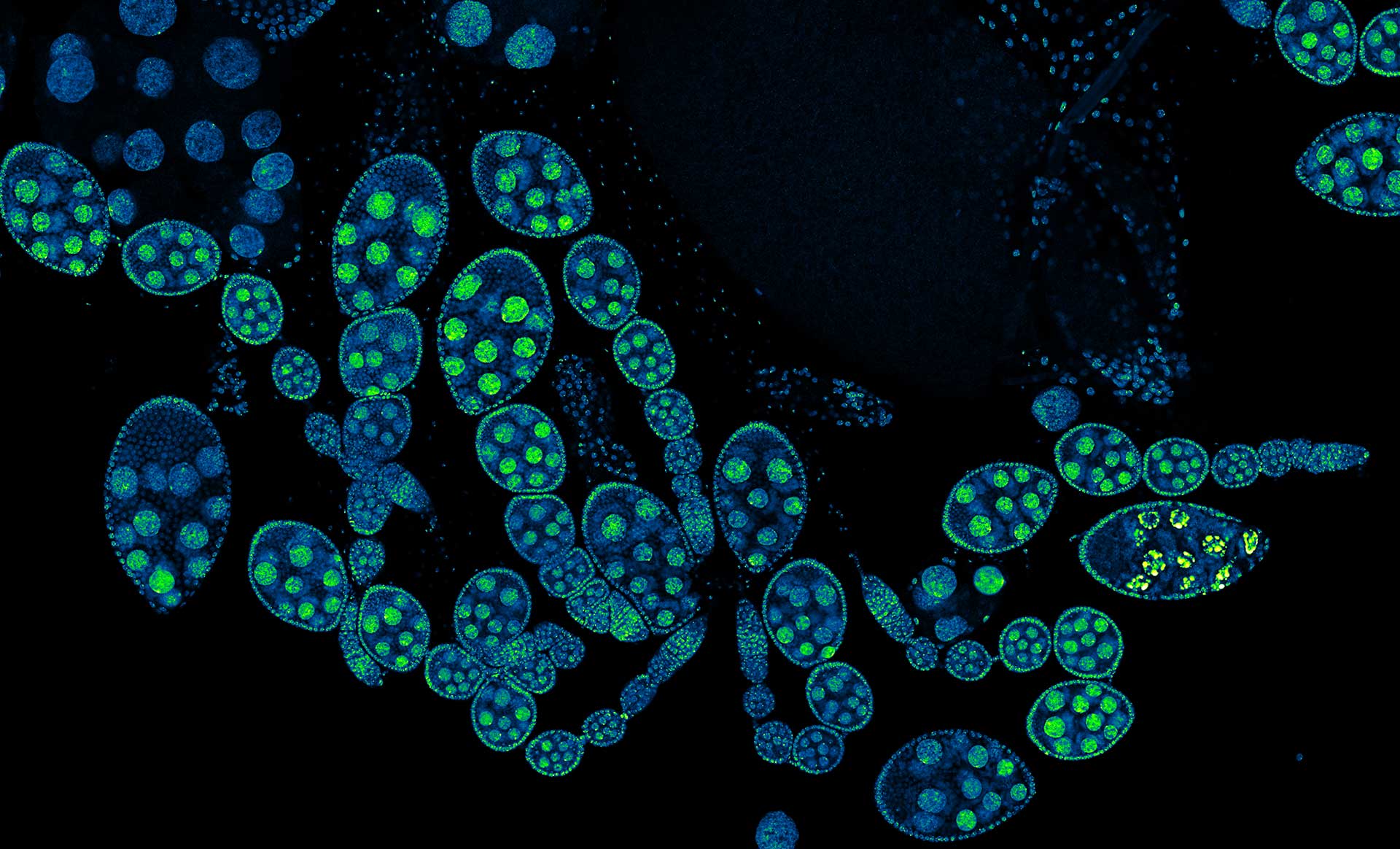
Description
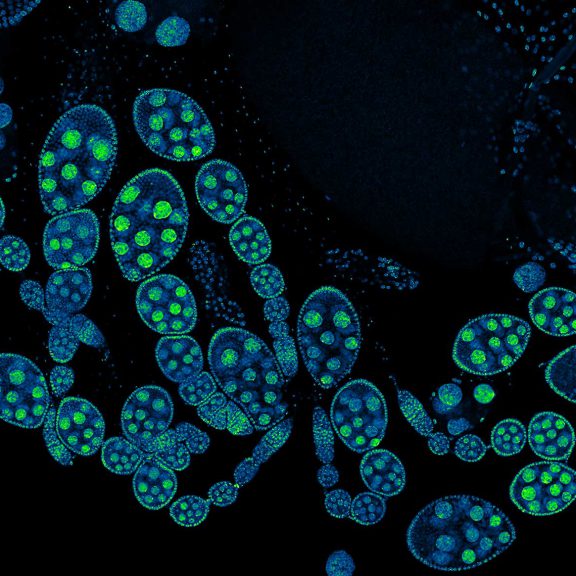
Drosophila ovariole stained with abberior LIVE 560 DNA showing nuclei in different cell types of the egg chamber. Ovaries were dissected from adult female fruit flies and were fixed prior to staining.
Image was acquired with the STEDYCON tiling feature and assembled with the SVI Huygens Stitcher.

Description
Drosophila spermatid tails were stained with abberior LIVE 610 tubulin. Testis were dissected from adult male fruit flies. Live cell imaging experiment was performed on a INFINITY microscope.
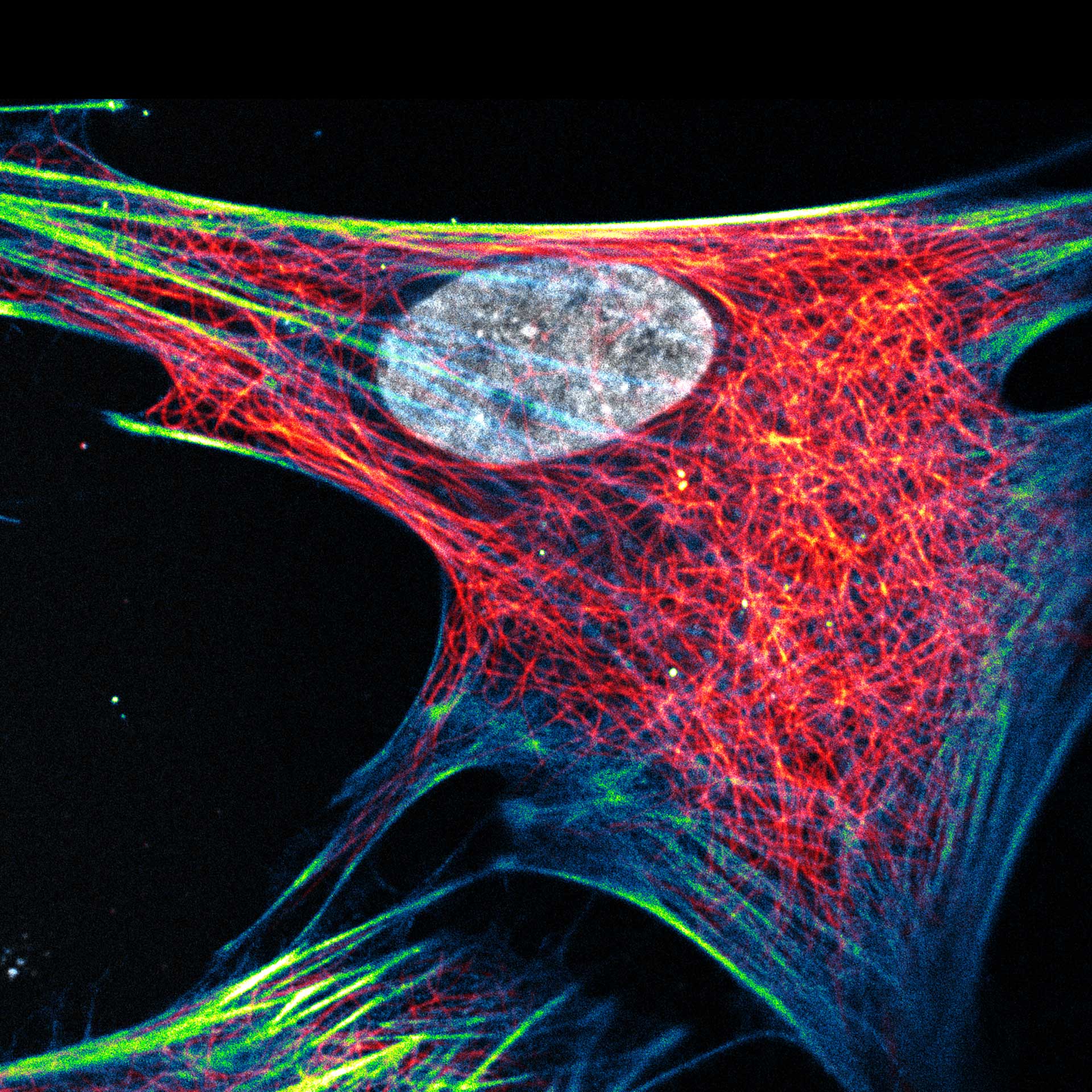
Description
Composite of three color live-cell image of an adherent mammalian cell. This living cell was directly labelled with abberior LIVE 510 actin (blue/green), LIVE 560 DNA (gray) and LIVE 610 tubulin. This image was acquired with a STEDYCON microscope.
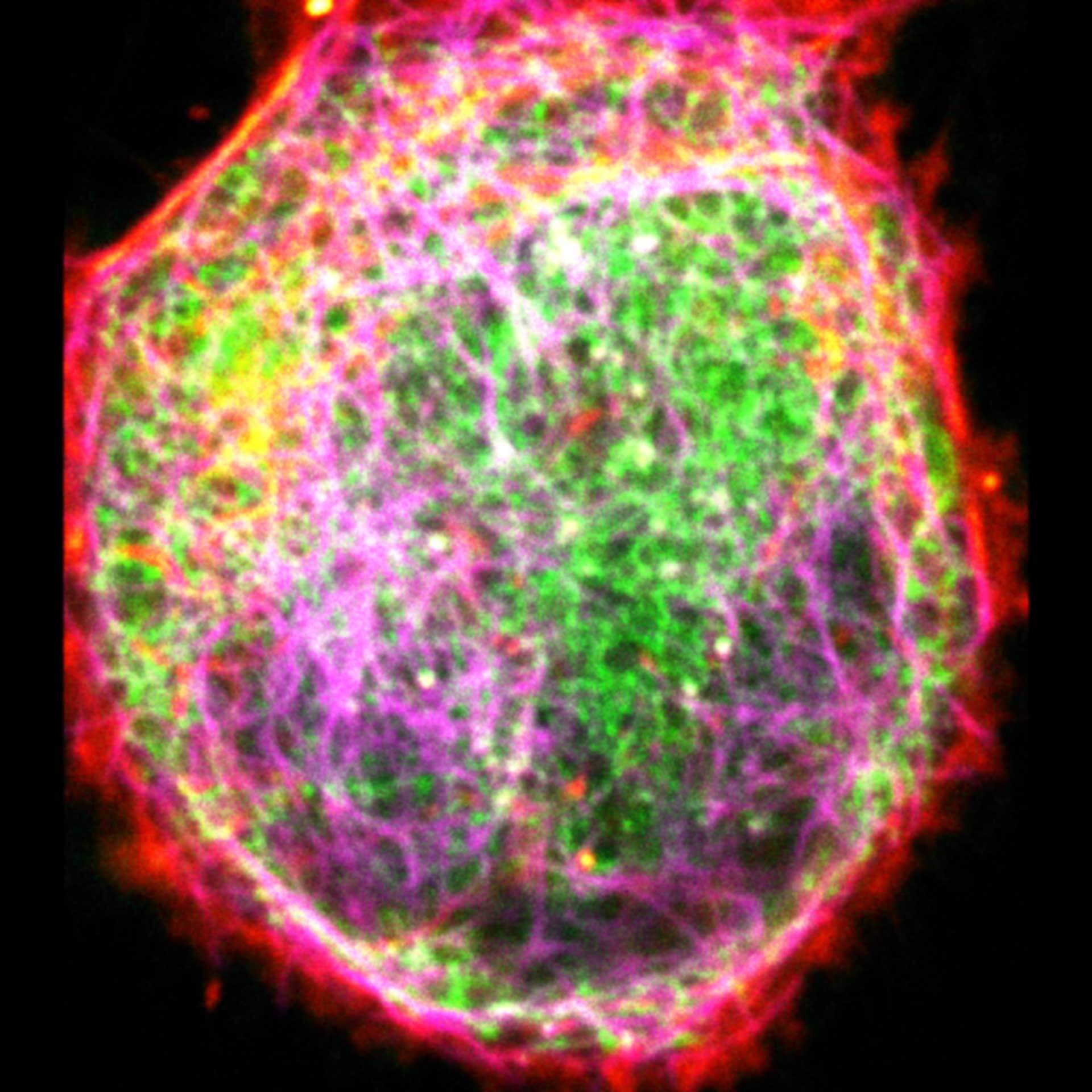
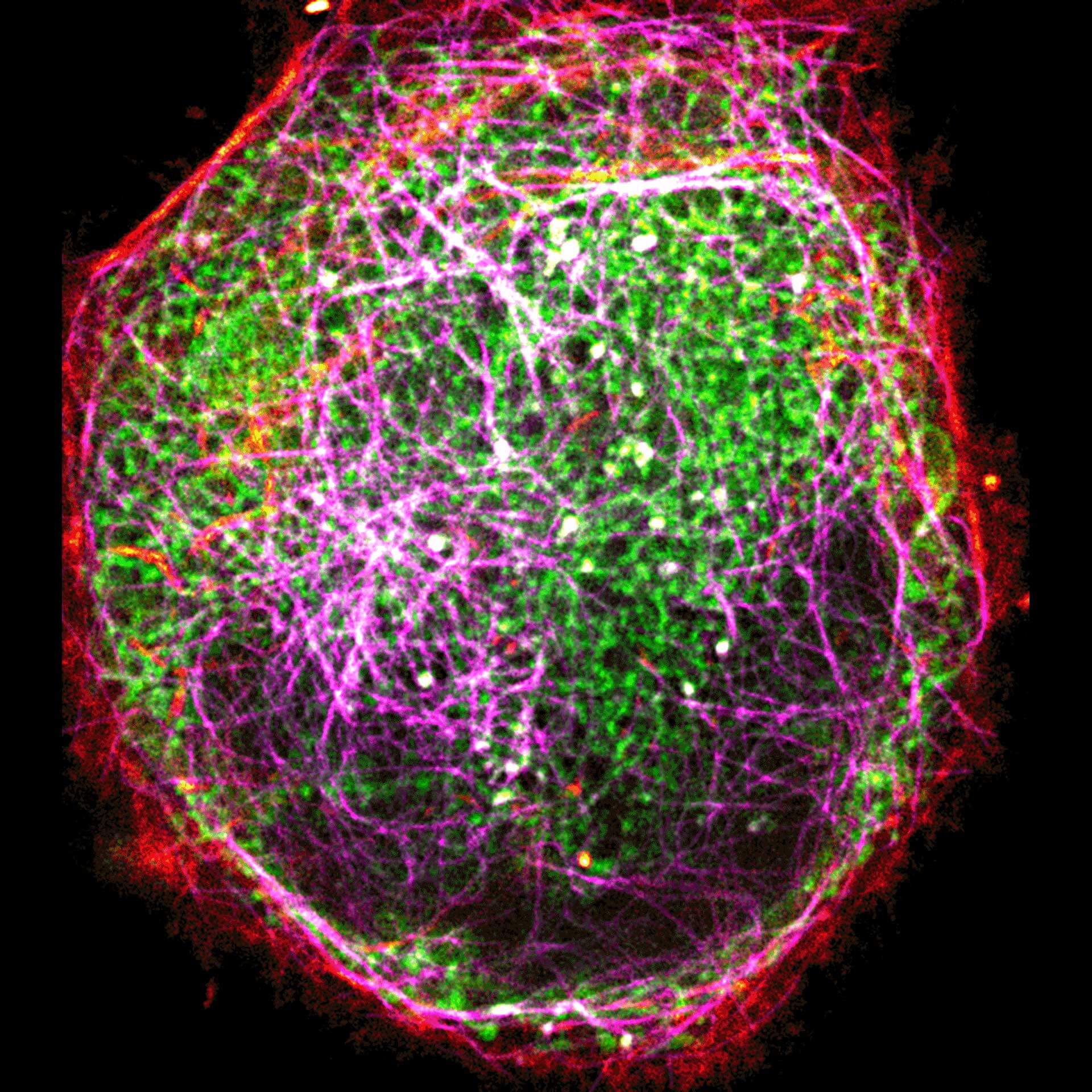
Description
Three color live-cell STED at 775 nm: living cell labelled with abberior LIVE 460L (ER, green), LIVE 560 tubulin (magenta) and LIVE 610 actin (red).
Description
Confocal and STED time lapse of a living cell expressing SNAP-tag® fusion protein fusion protein in the endoplasmic reticulum lumen with SNAP and C-terminal tetrapeptide KDEL. SNAP-KDEL was labelled with abberior LIVE 610 SNAP ligand (benzylguanine).
The SNAP-KDEL plasmid was a gift from Francesca Bottanelli, FU Berlin.
Description
Live cell time lapse of abberior LIVE 590 DNA probe in cultured mammalian cells.
The use of very low concentrations of our abberior LIVE dyes reduces toxicity and allows long-term imaging of dynamic processes.
Image was acquired with the FACILITY microscope.
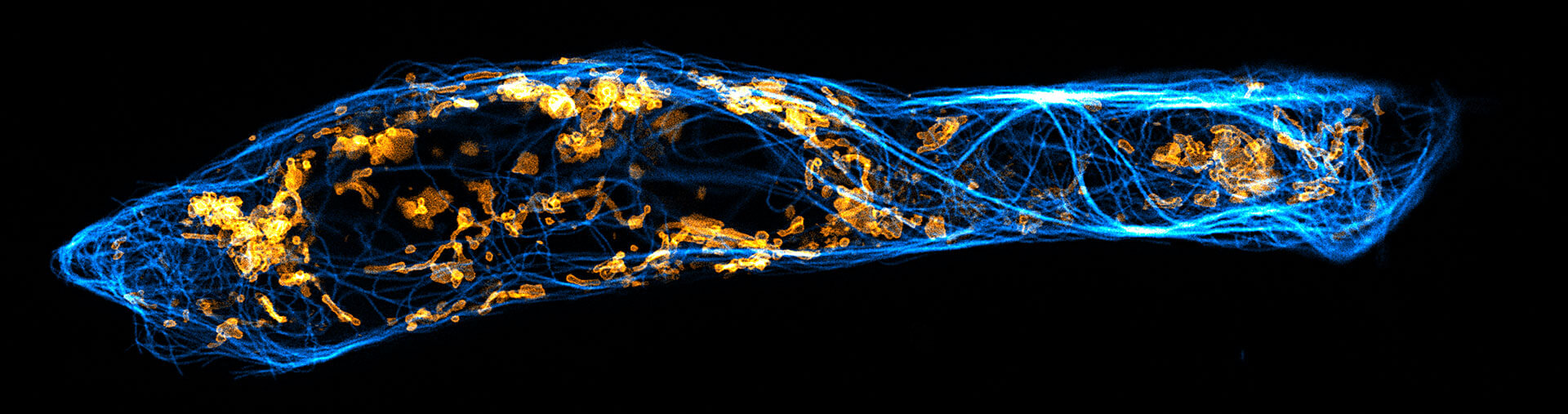
Description
Two color live-cell confocal and STED image of a mammalian cell expressing a SNAP-tag® OMP25 fusion protein decoration the outer membrane of mitochondria. OMP25 is visualized by our new abberior LIVE 610 SNAP ligand (orange). Tubulin filaments are highlighted with abberior LIVE 550 tubulin (cyan).
The SNAP-OMP25 plasmid was a gift from Francesca Bottanelli, FU Berlin.
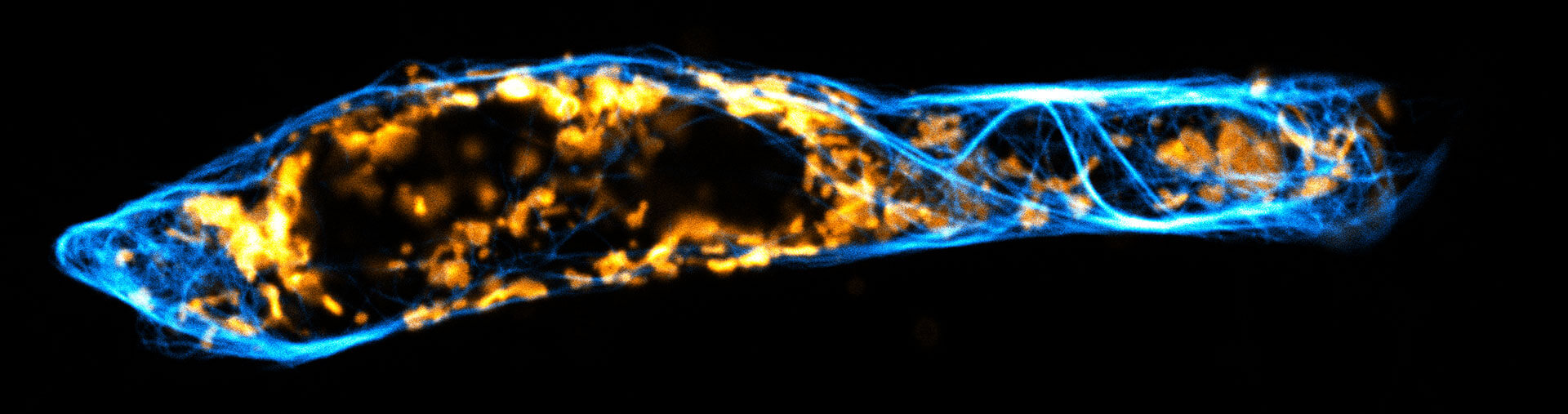

Description
Two color live-cell confocal and STED image of a mammalian cell expressing a SNAP-tag® OMP25 fusion protein decoration the outer membrane of mitochondria. OMP25 is visualized by our new abberior LIVE 610 SNAP ligand (orange). Tubulin filaments are highlighted with abberior LIVE 550 tubulin (cyan).
The SNAP-OMP25 plasmid was a gift from Francesca Bottanelli, FU Berlin.
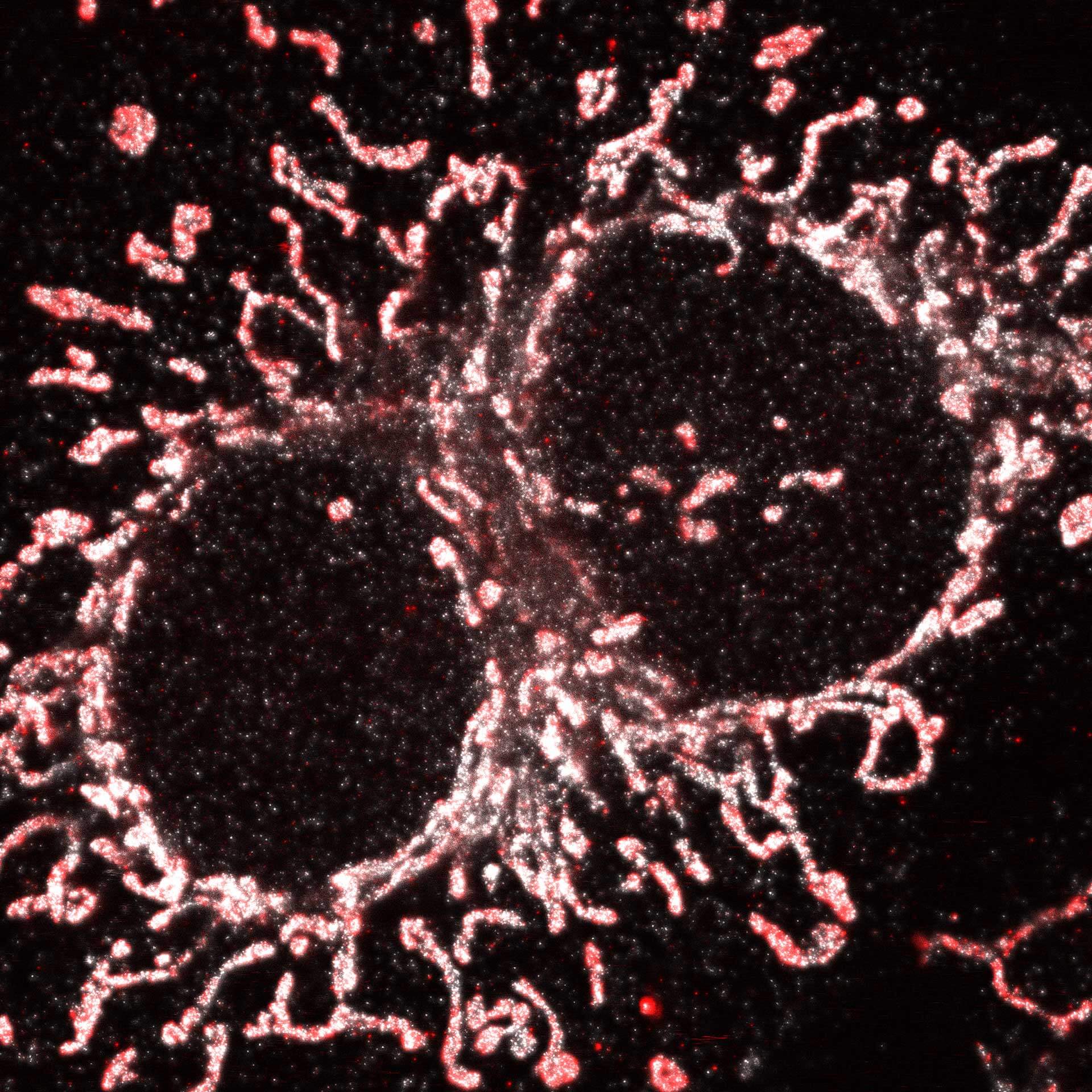
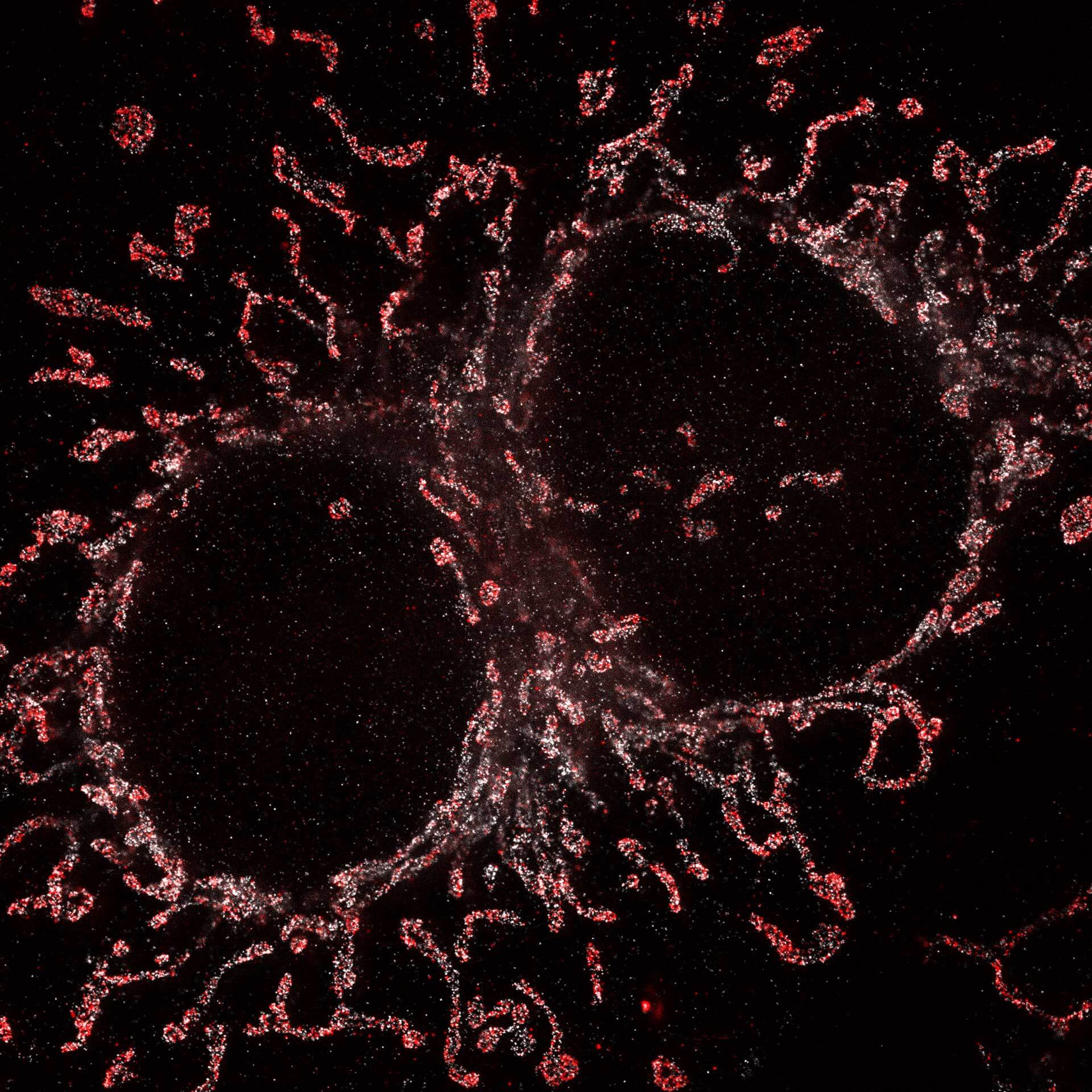
Description
Cultured mammalian cell immunostained for an inner and outer mitochondrial membrane marker. Outer membrane is highlighted with abberior STAR RED (red) and the inner membrane with abberior STAR ORANGE (gray).
STED and confocal images were acquired with abberior's FACILITY microscope.
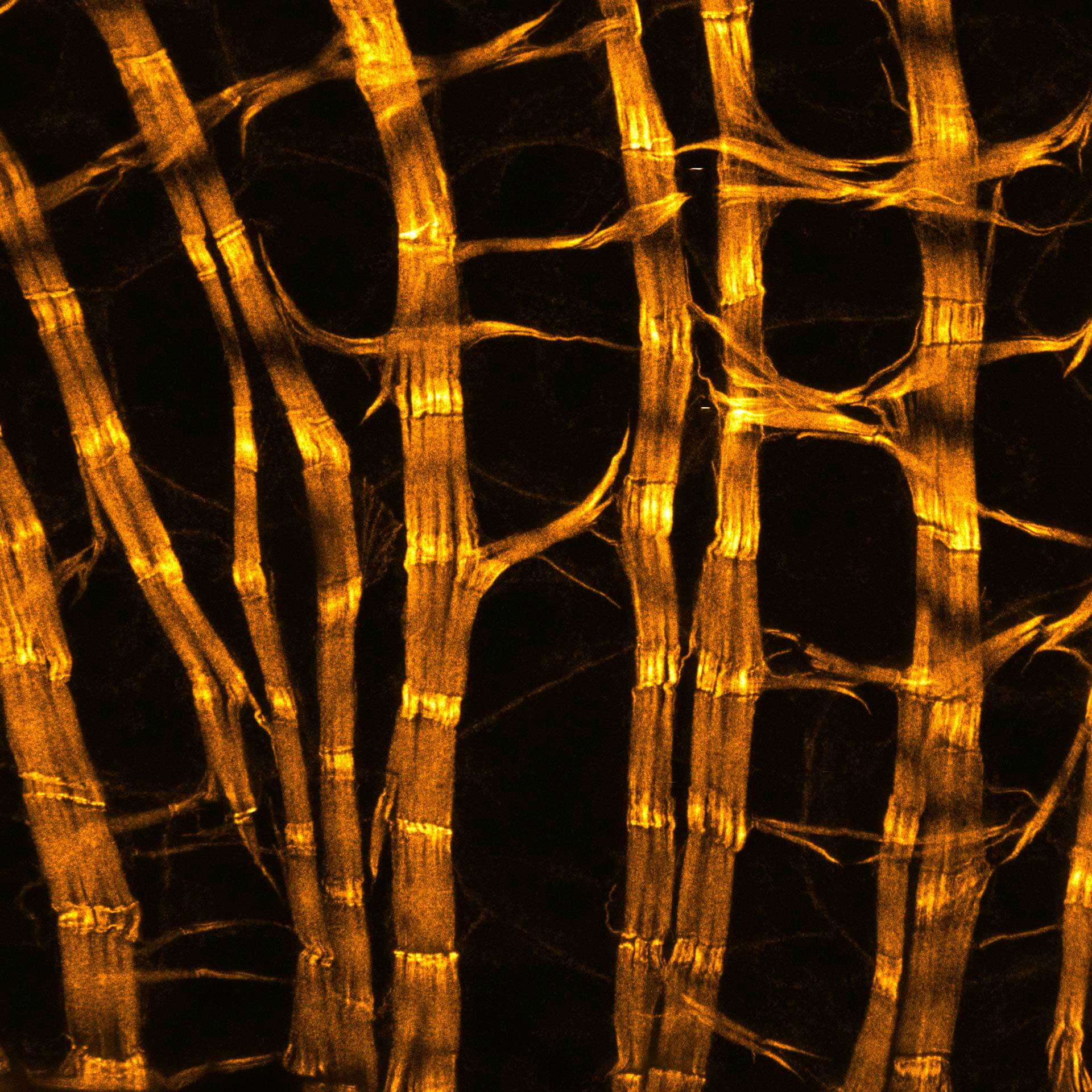
Description
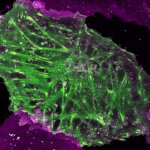
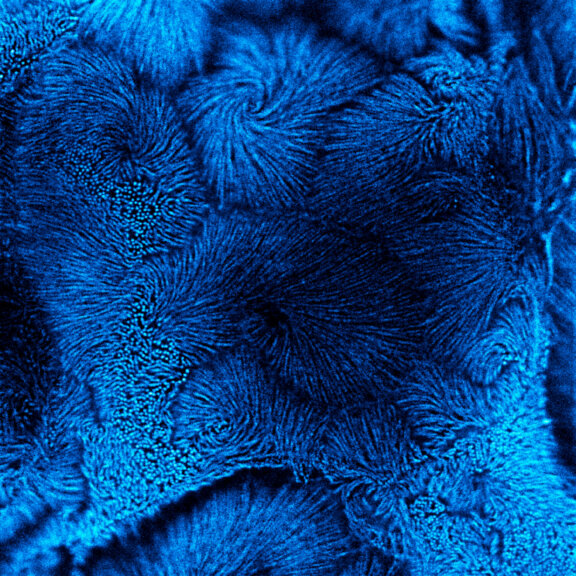
Drosophila male accessory gland stained for F-actin using abberior STAR 580 phalloidin.
Sample was prepared in cooperation with Dr. H. R. Shcherbata at MPl for Biophysical Chemistry, Göttingen, Germany.
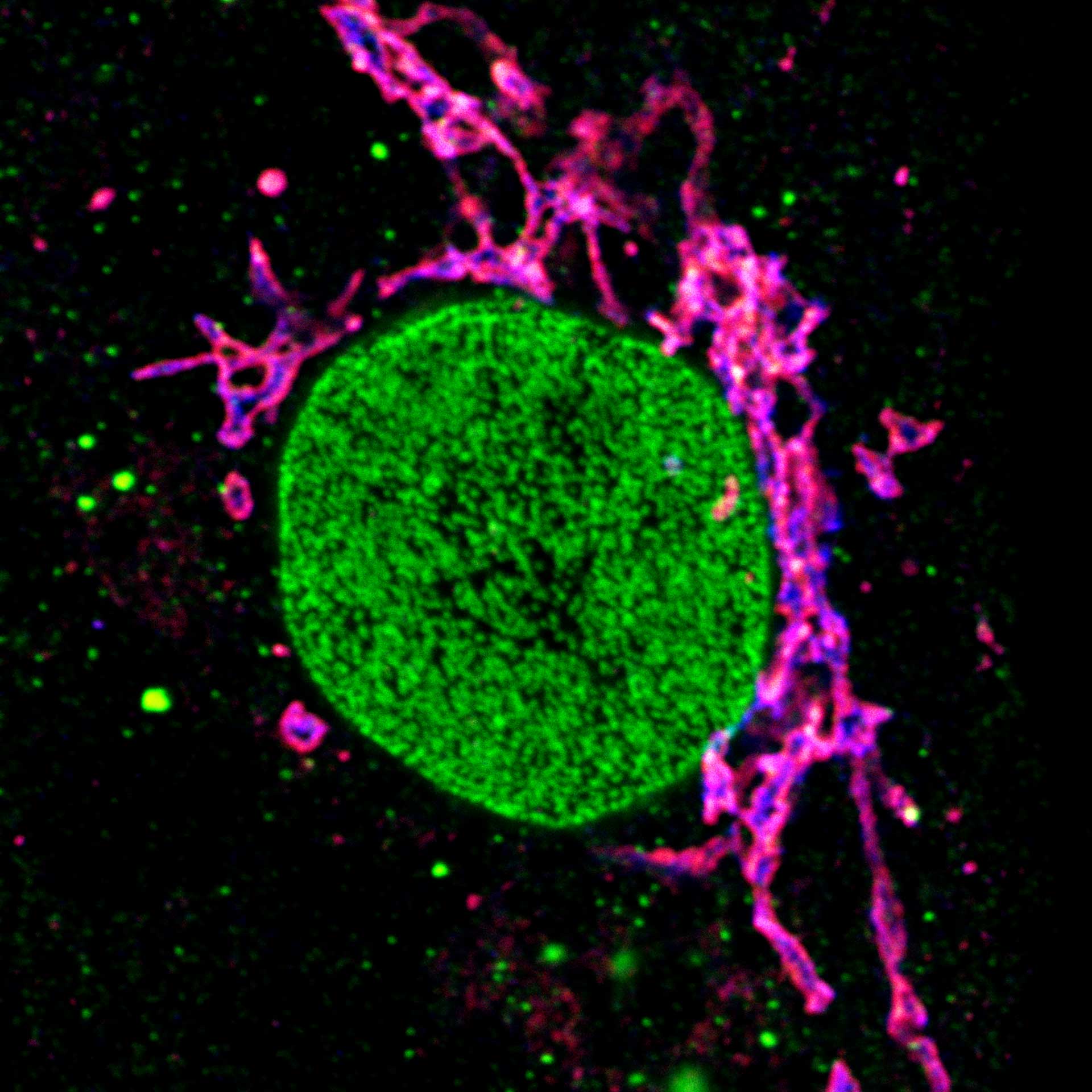
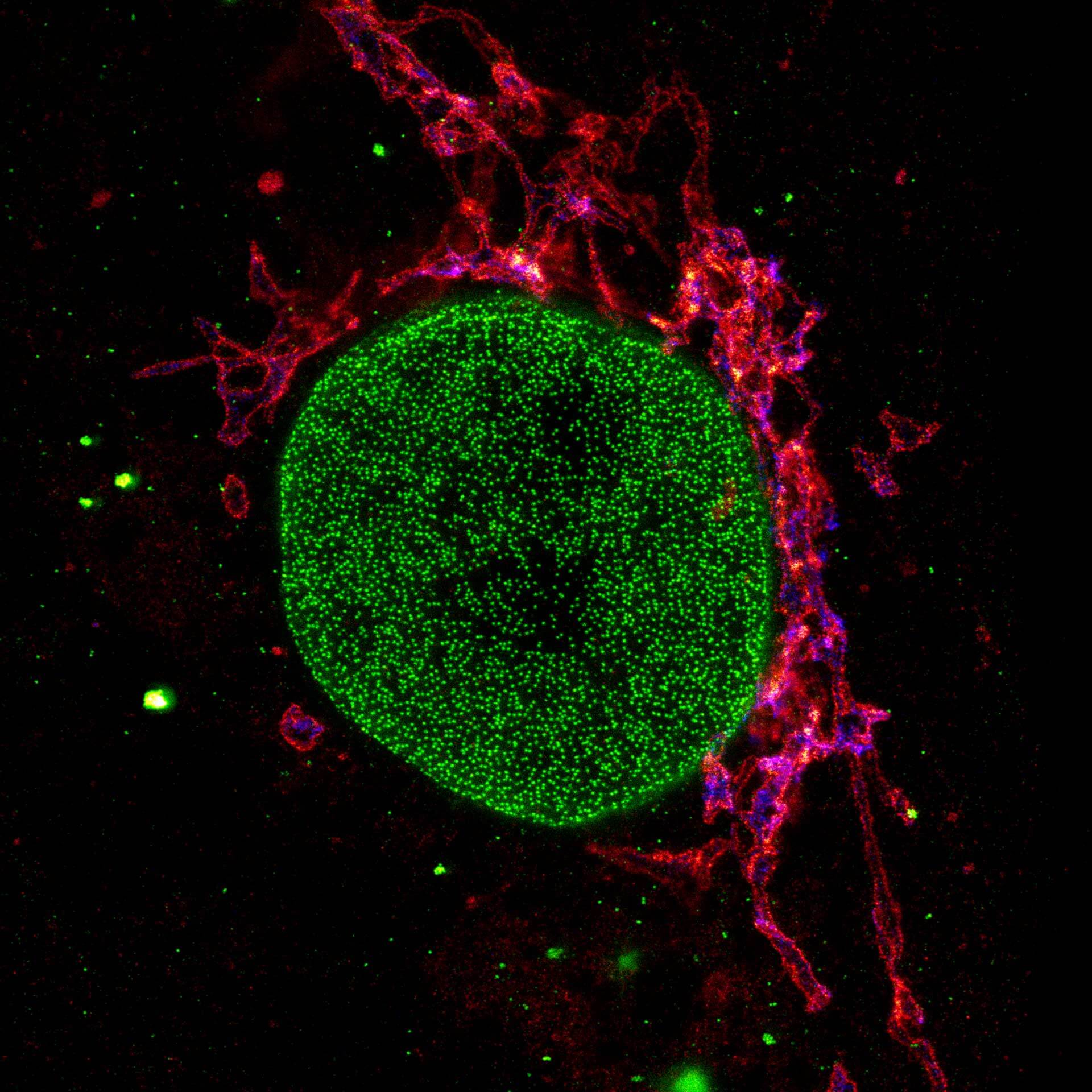
Description
Three color STED and confocal image of a mammalian cultured cell immunostained for a nuclear pore protein in green and two golgi apparatus markers in red and blue.
abberior STAR RED highlights the golgi apparatus protein giantin (red) and abberior STAR ORANGE visualizes the cis-golgi protein GM130 (blue). NUP98 proteins were stained with abberior STAR GREEN (green).
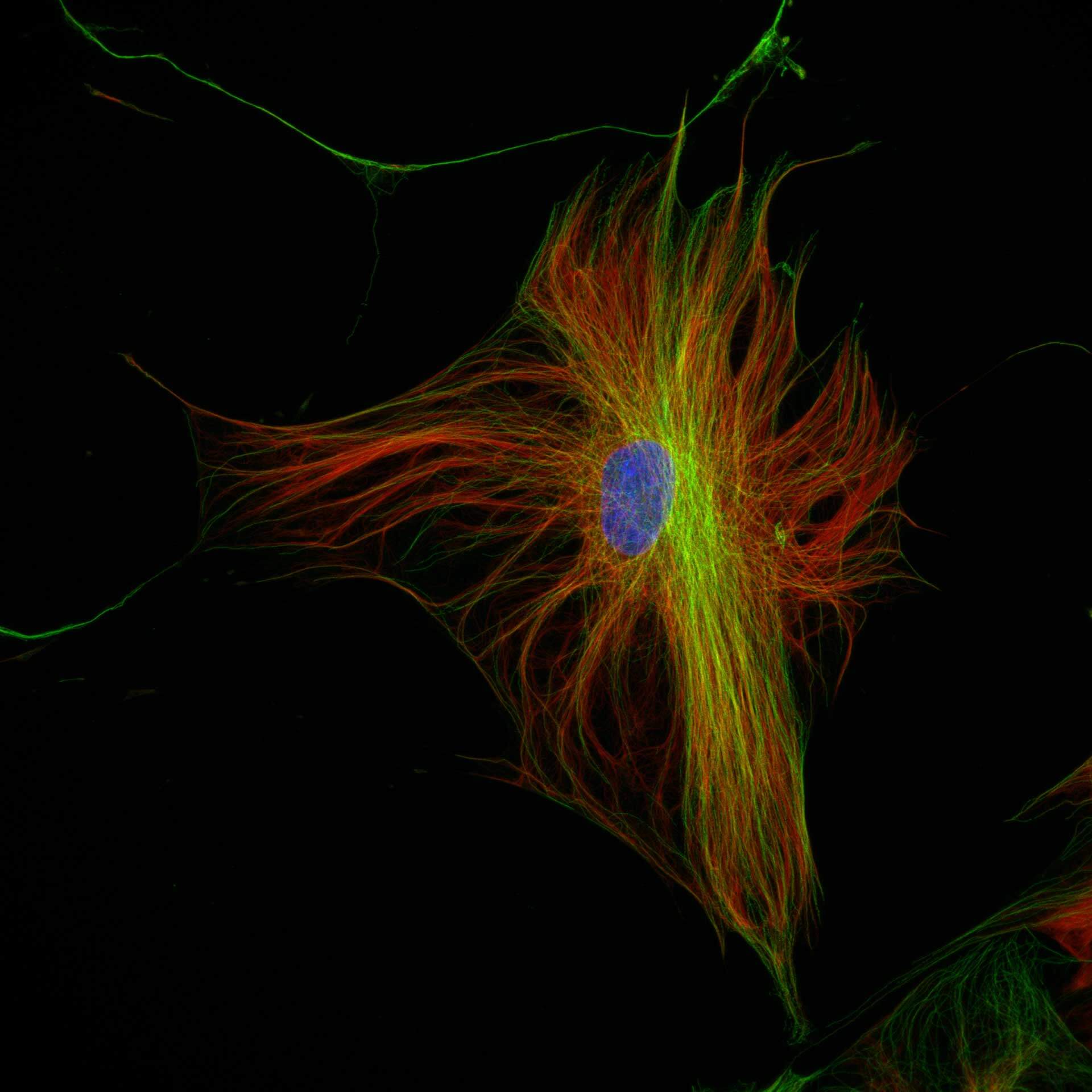
Description
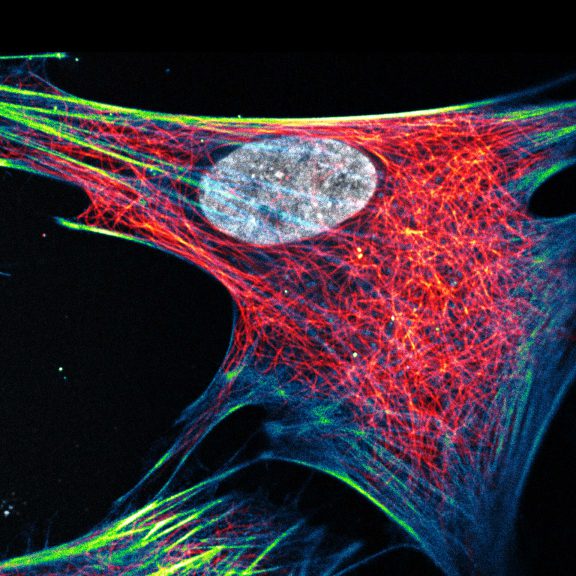
Three color confocal image of human fibroblast immunostained with abberior STAR 580 for vimentin (green) and with abberior STAR RED for tubulin (red).
DAPI was added to highlight the nucleus (blue).
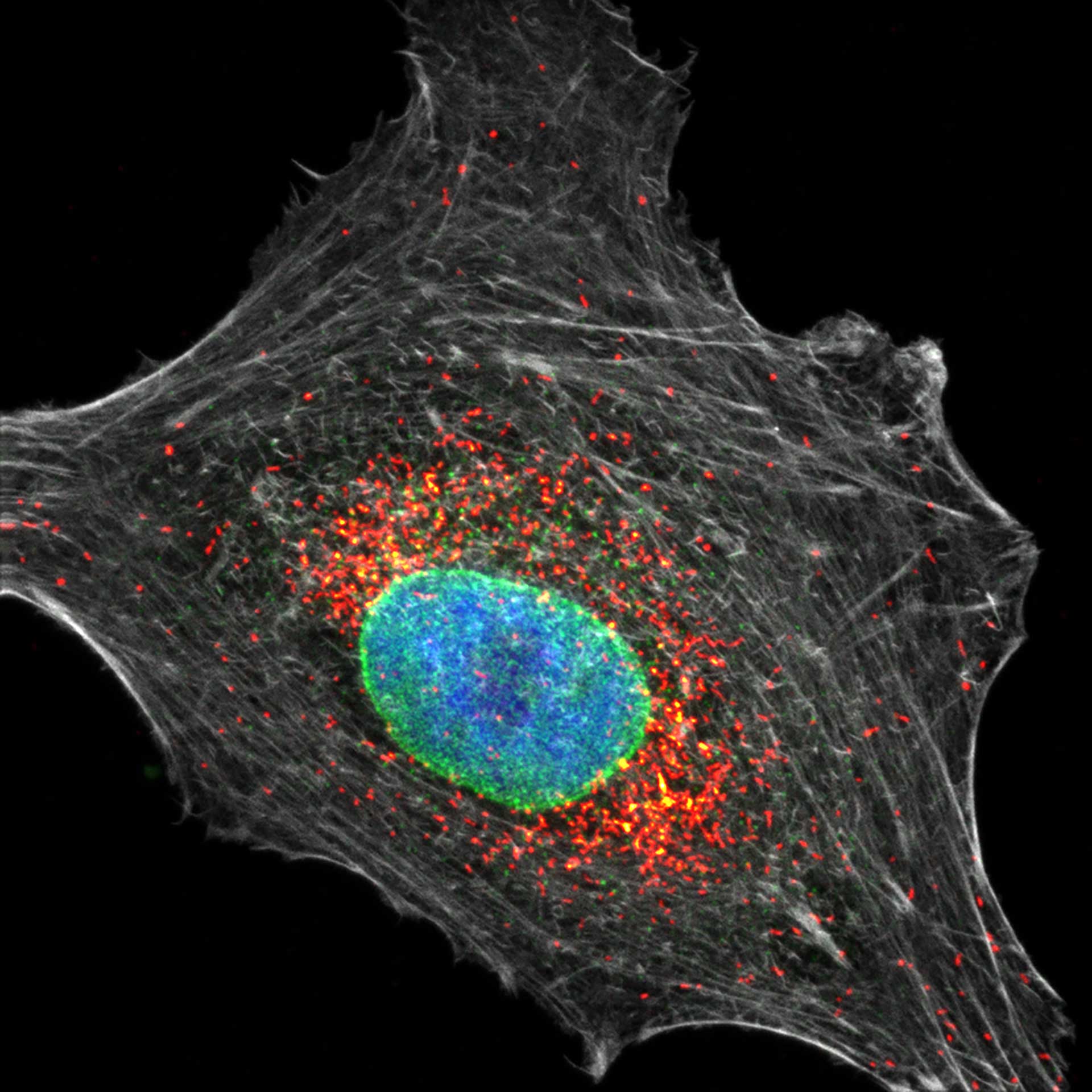
Description
Four color confocal image of mammalian cultured cells. Peroxisomes were immunostained with abberior STAR RED (magenta) and nuclear pore proteins with abberior STAR 580 (green). F-actin fibers were highlighted with abberior STAR 488 phalloidin (gray).
The nucleus was visualized with DAPI (blue).
Description
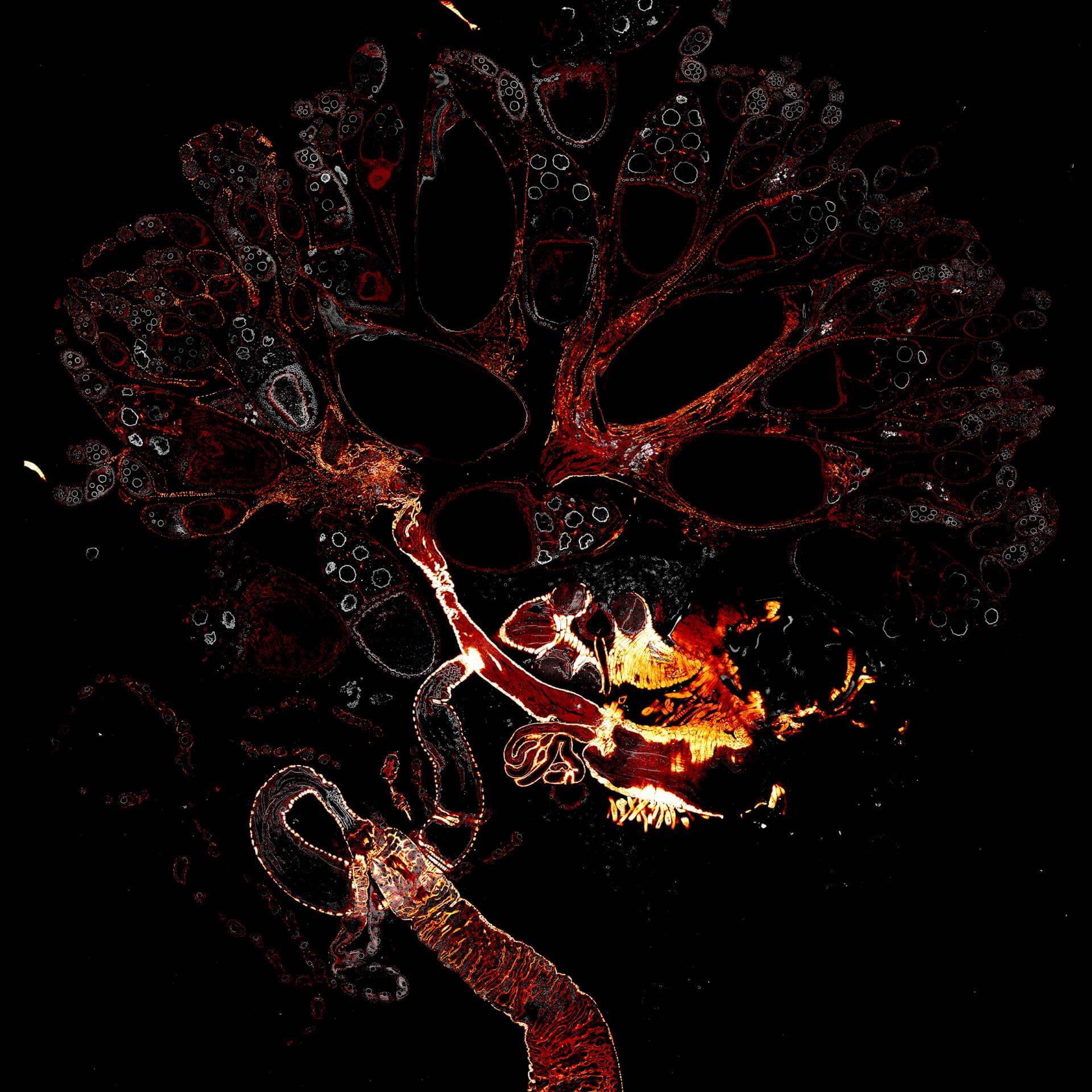
Drosophila female reproductive system stained for F-actin (red) with abberior STAR RED phalloidin. abberior STAR ORANGE is highlighting a nuclear pore protein (gray).
Image was acquired with the STEDYCON tiling feature and assembled with the SVI Huygens Stitcher.
Description
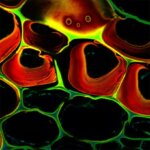
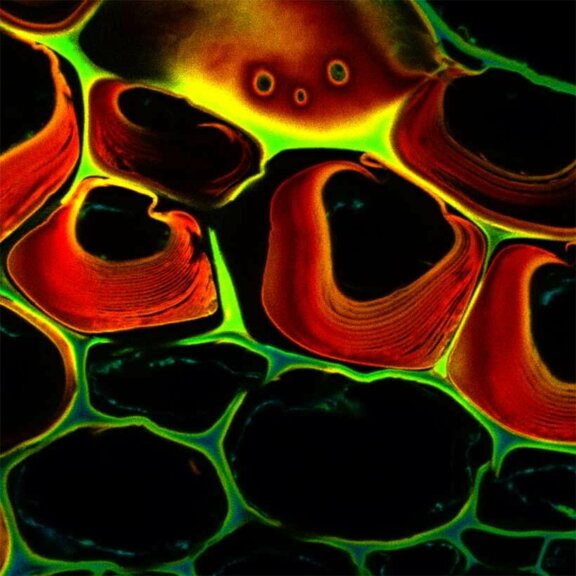
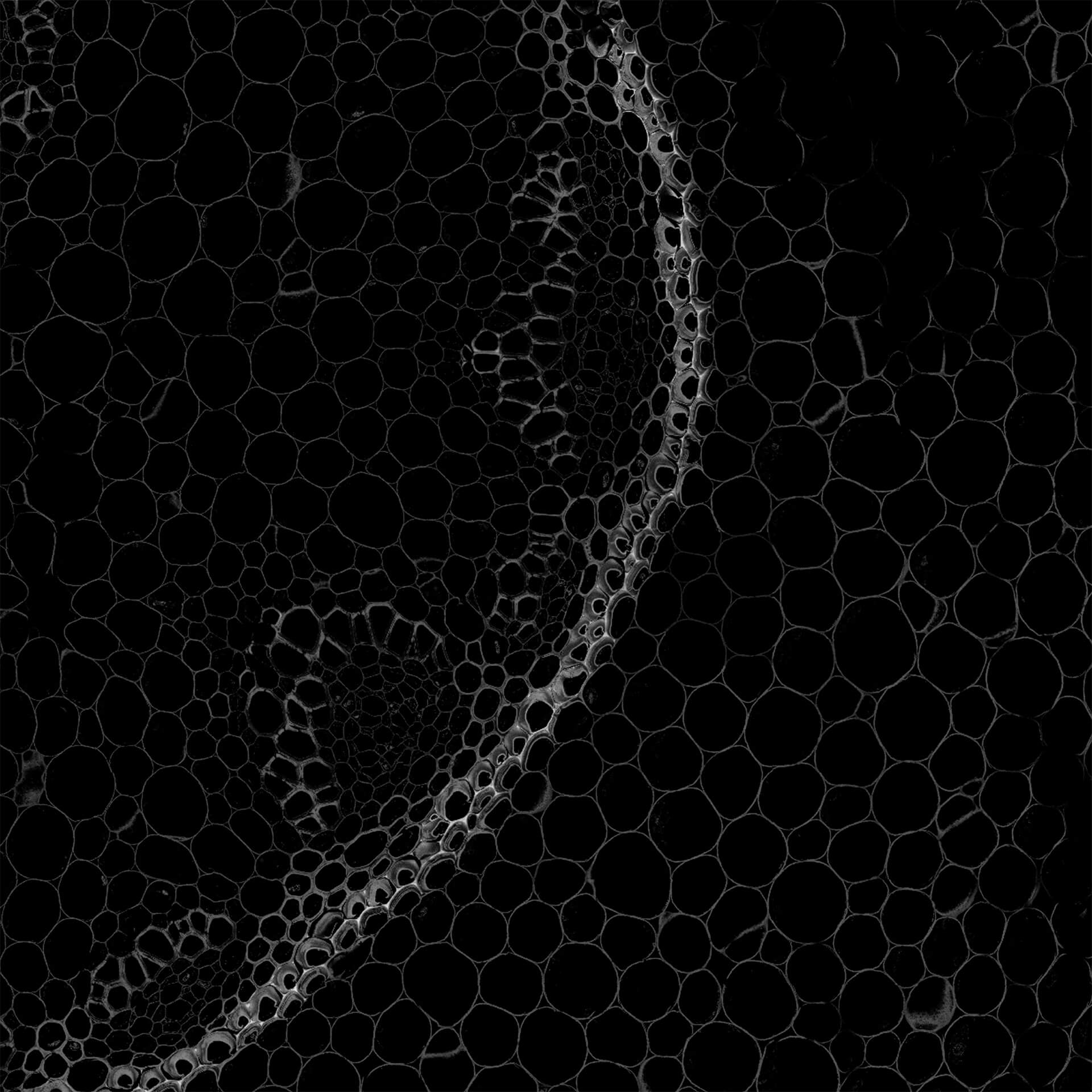
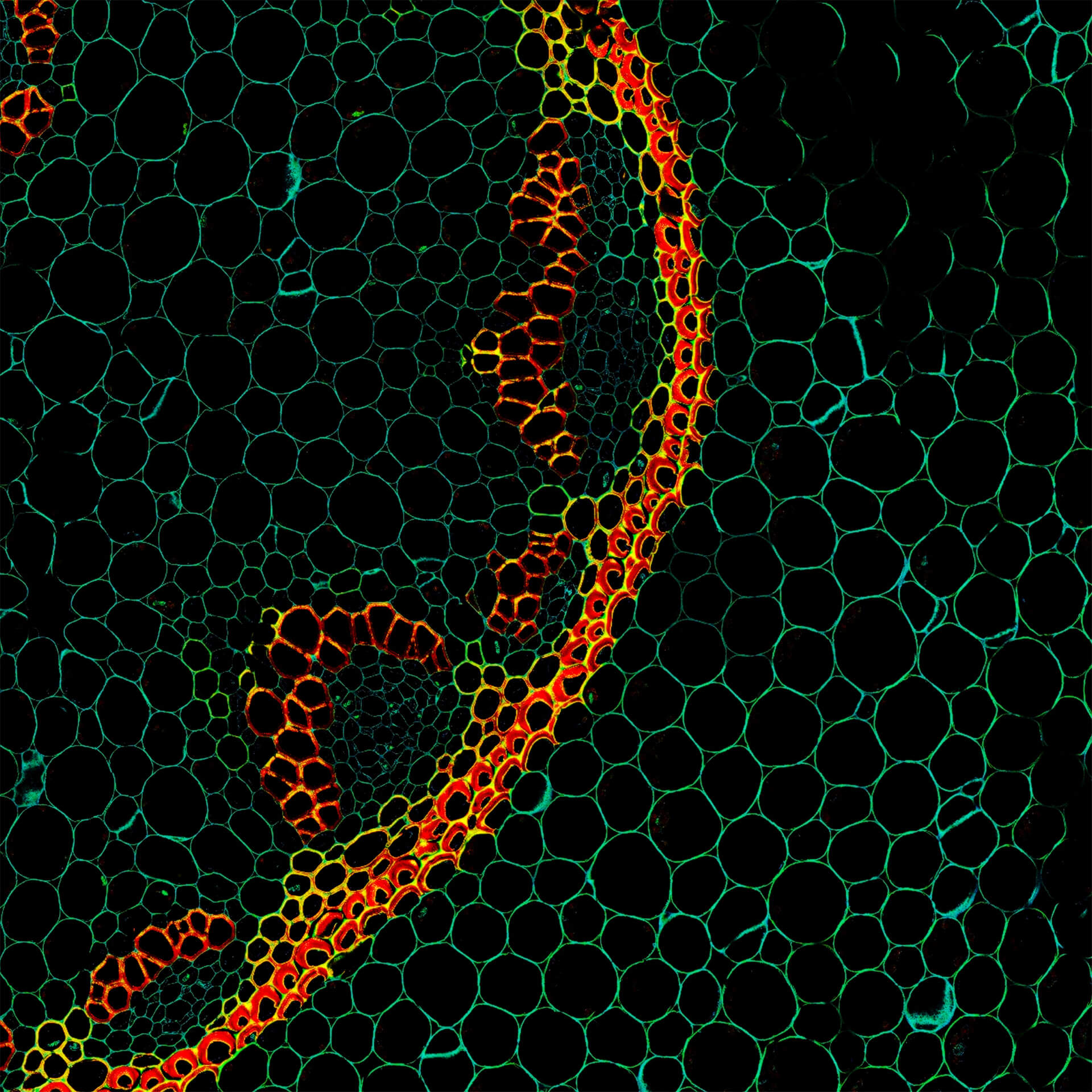
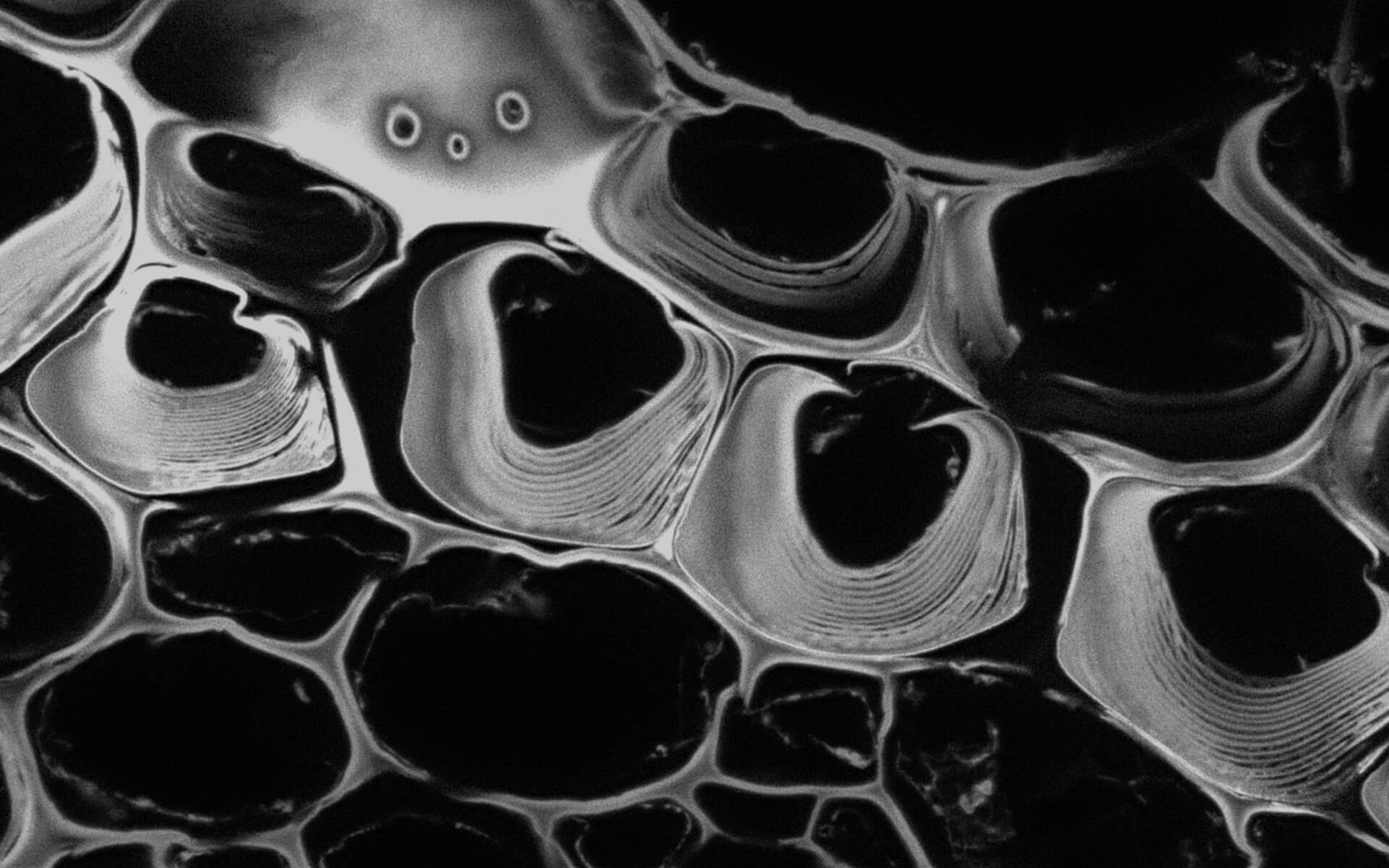
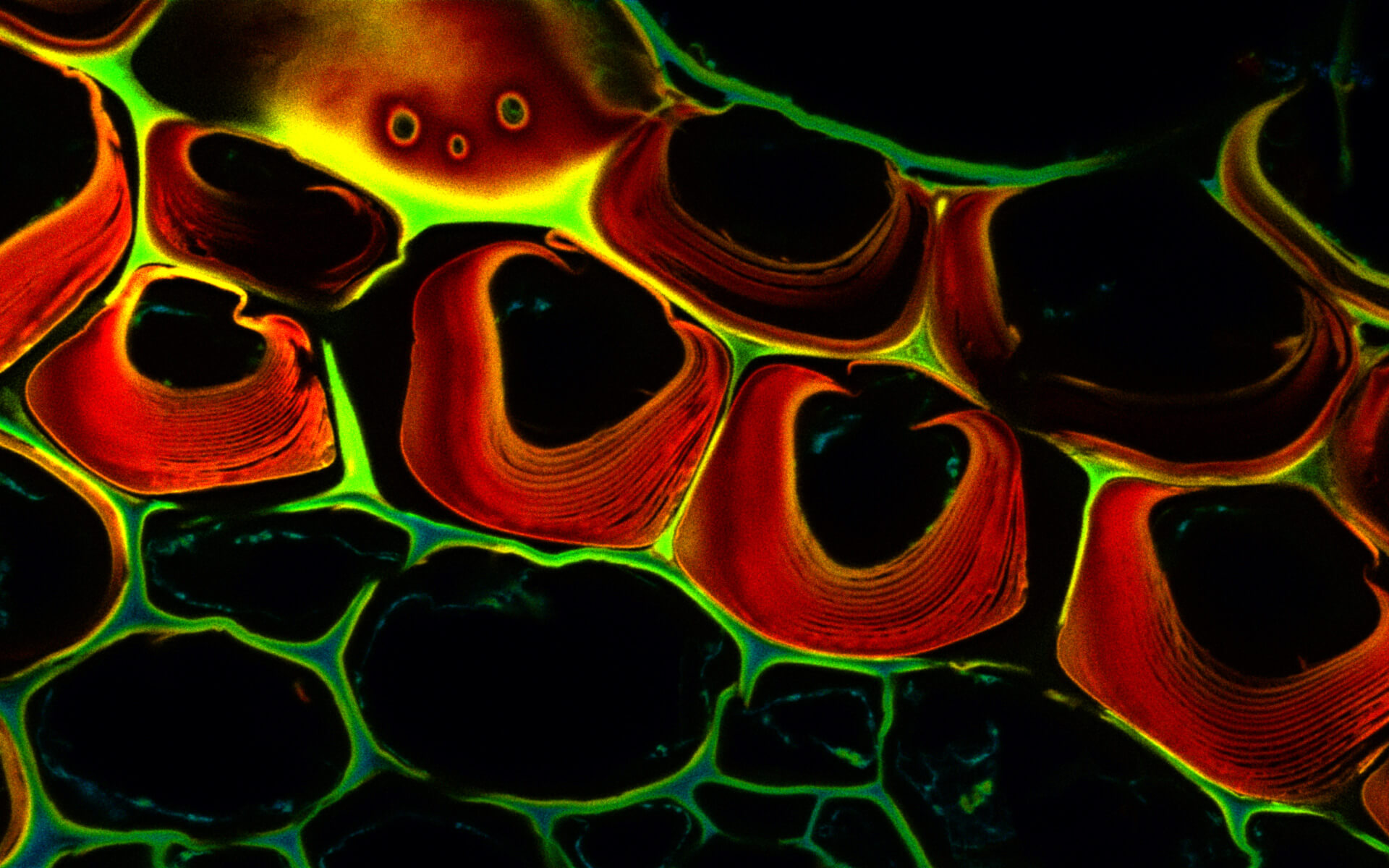
Autofluorescence in a plant stem cross section of Convallaria recorded with FACILITY. A large area, 10 by 10 tiles, each 125 µm by 125 µm, was imaged and stitched with image tiling in Fiji for ImageJ. This is combined with TIMEBOW lifetime imaging to show the differences in fluorescence lifetime due to the type of autofluorescent molecule and their nano-environment.
Modules:
Description
Live-cell sample of an Arabidopsis root tip suspended in water recorded with FACILITY. A subset of cells expresses a YFP construct. Lifetime imaging with TIMEBOW shows shifts in YFP fluorescence lifetime caused by the proteins nano-environment.
We thank Dr. Fábián Attila and Soós Vilmos (ATK, Brunszvik, HU) for providing this sample. Contact: soos.vilmos@atk.hu.
Modules:
Description
Autofluorescence in a plant stem cross section of Convallaria recorded with FACILITY. Image quality is improved by removing the background through MATRIX detection. This is combined with TIMEBOW lifetime imaging to show the differences in fluorescence lifetime due to the type of autofluorescent molecule and their nano-environment.
Modules:

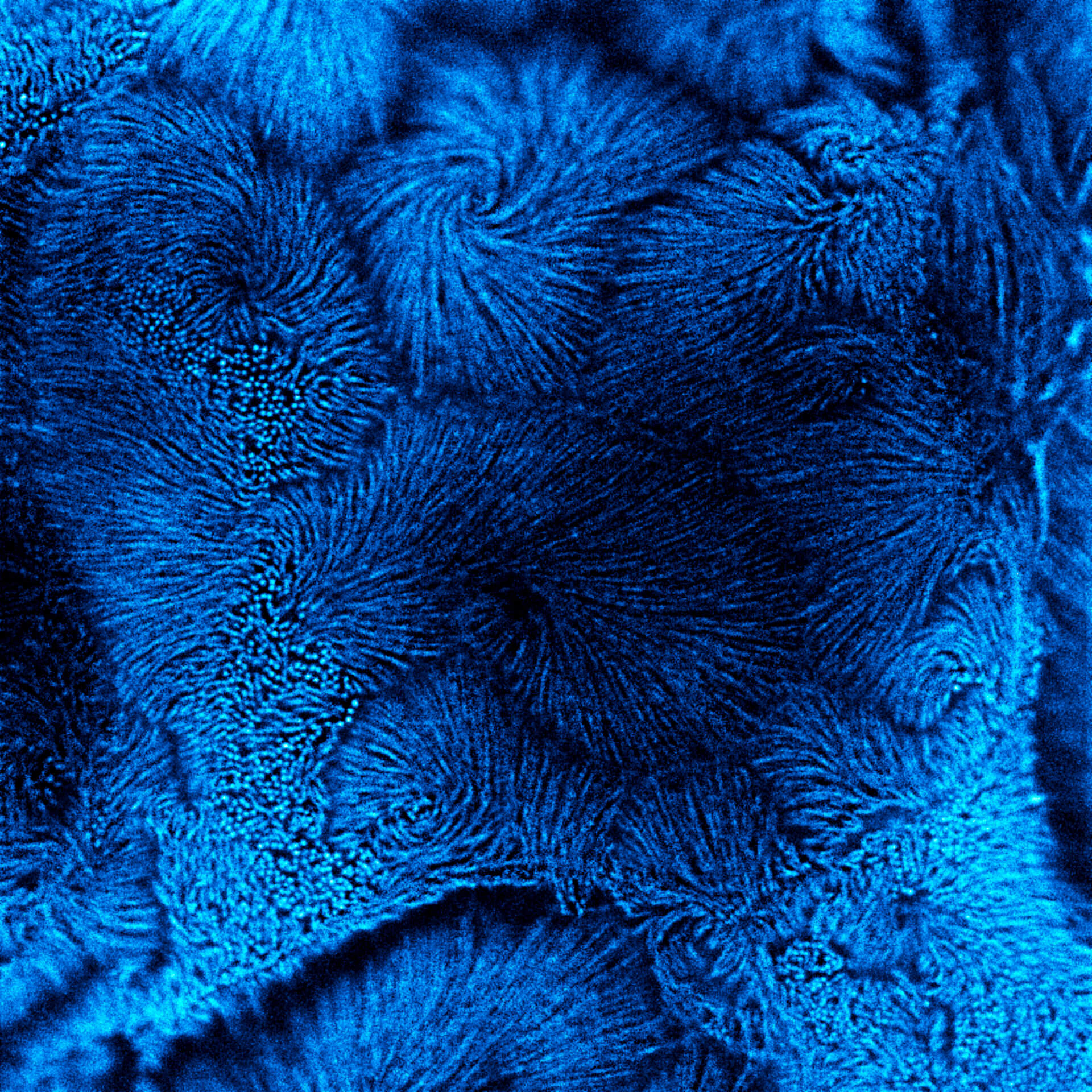
Description
TIMEBOW STED vs MATRIX + TIMEBOW STED
Actin in microvilli of CaCo2 cells recorded with FACILITY. Fixed cells were stained with abberior STAR RED phalloidin. Imaging of the intense-background sample is optimized by MATRIX detection by directly subtracting unfocussed data. To boost resolution TIMEBOW is used, keeping STED laser power low. Lifetime information enables the selective exclusion of STED-affected emission at the rim of each measurement.
We thank Prof. Dorothee Günzel and Jörg Piontek (Charité, Berlin, DE) for providing this sample.
Modules:
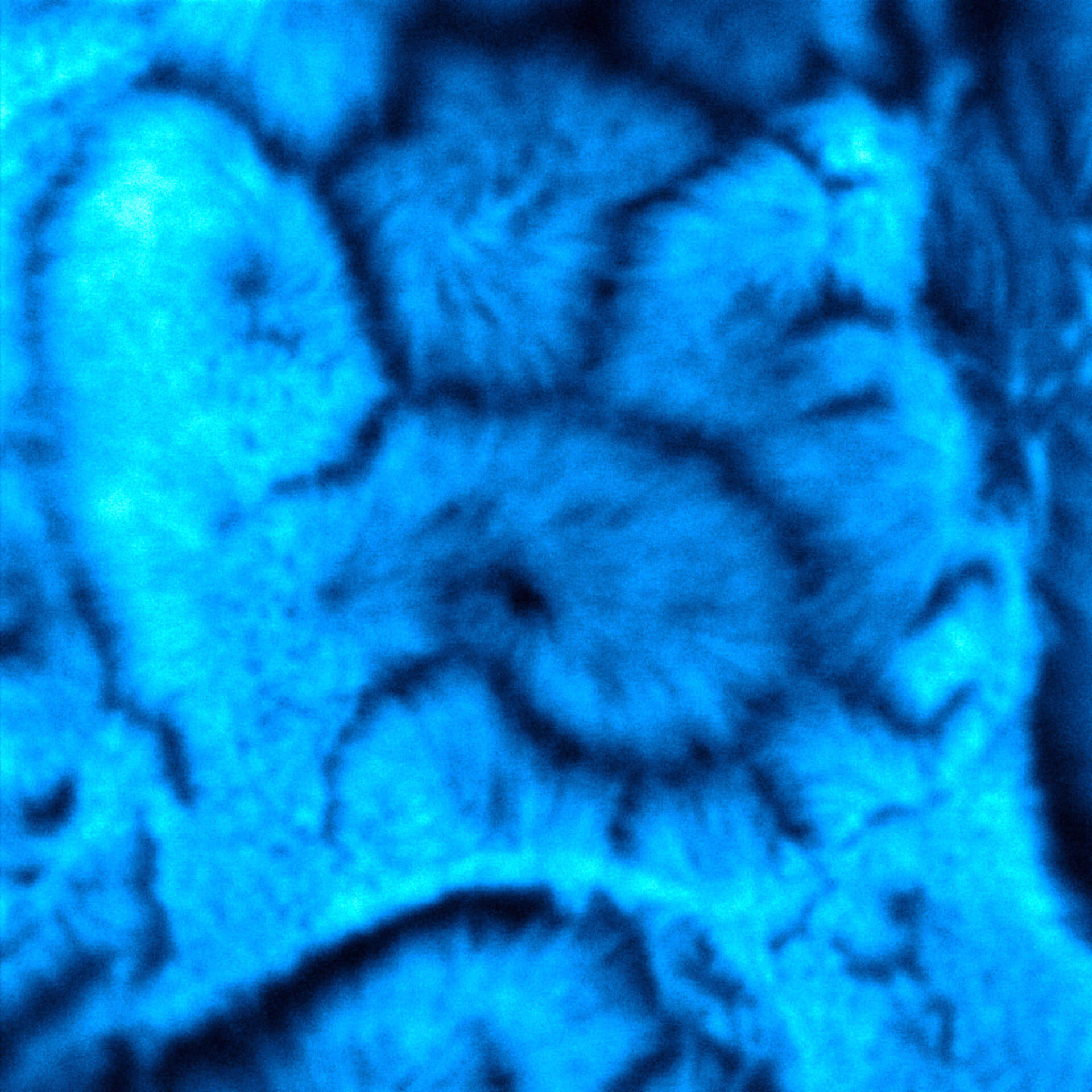

Description
Confocal vs MATRIX + TIMEBOW STED
Actin in microvilli of CaCo2 cells recorded with FACILITY. Fixed cells were stained with abberior STAR RED phalloidin. Imaging of the intense-background sample is optimized by MATRIX detection by directly subtracting unfocussed data. To boost resolution TIMEBOW is used, keeping STED laser power low. Lifetime information enables the selective exclusion of STED-affected emission at the rim of each measurement.
We thank Prof. Dorothee Günzel and Jörg Piontek (Charité, Berlin, DE) for providing this sample.
Modules:
Description
MINFLUX 3D imaging of βII spectrin in a primary hippocampal neuron labeled with Alexa Fluor 647 by indirect immunofluorescence. The axial coordinate is color-coded. Please note the periodic arrangement of spectrin along the axon.
Description
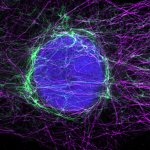
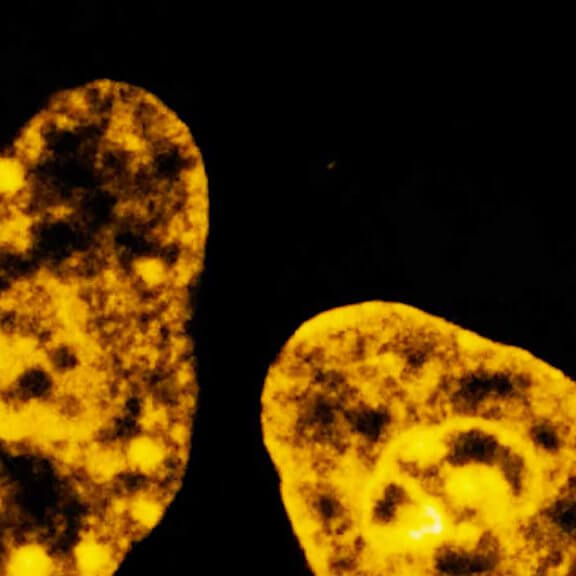
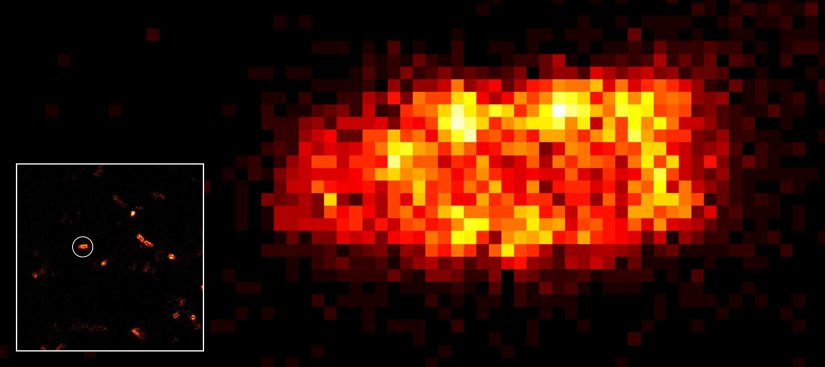
MINFLUX 3D nanoscopy of nuclear pore complex subunits. 3D MINFLUX facilitates 3D imaging at molecular scales. In the axial direction localization, we reach precisions of approx. 2.5 nm in raw localization data. Labeling of NUP96-SNAP was performed using SNAP-Alexa Fluor 647.
Description
3D MINFLUX imaging of the peroxisomal membrane protein PMP70 labelled with abberior FLUX 647 in fixed mammalian cells using indirect immunofluorescence. 3D MINFLUX allows visualizing the shape of peroxisomes in all directions.
Description
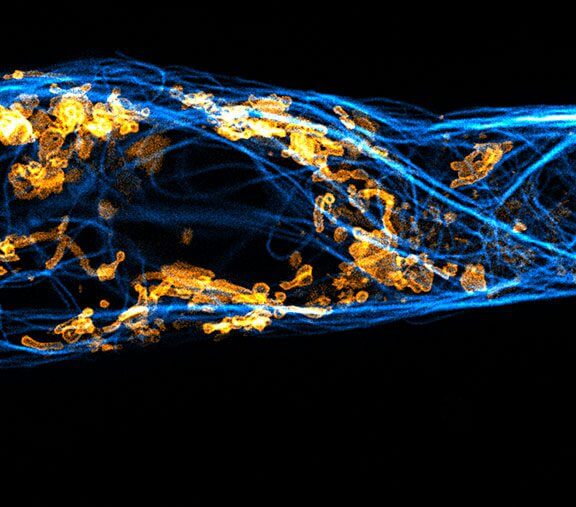
MINFLUX 3D imaging of Clathrin coated vesicles and Clathrin coated pits. Labeling was performed using clathrin light chain-SNAP together with SNAP-Alexa Fluor 647.
Description
2D MINFLUX nanoscopy of Y. enterocolitica sorting platform protein Halo-YscL.
Sample from Alexander Carsten, Prof. Martin Aepfelbacher, Institute of Medical Microbiology, Virology and Hygiene, University Medical Center Hamburg Eppendorf, Germany
Description
STED-PAINT imaging of bacterial membranes (magenta) and DNA (green) using a high concentration of the exchangeable labels.
Description
3D MINFLUX nanoscopy of Y. enterocolitica sorting platform protein Halo-YscL in xy view of two bacteria. The z-axis is color coded.
Sample from Alexander Carsten, Prof. Martin Aepfelbacher, Institute of Medical Microbiology, Virology and Hygiene, University Medical Center Hamburg Eppendorf, Germany
Description
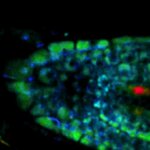
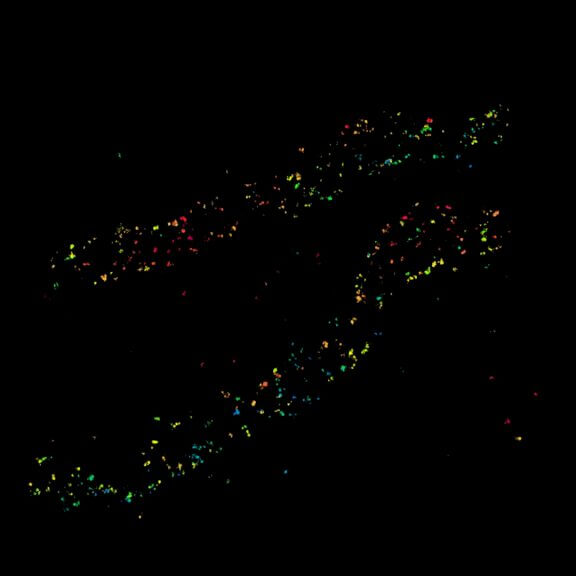
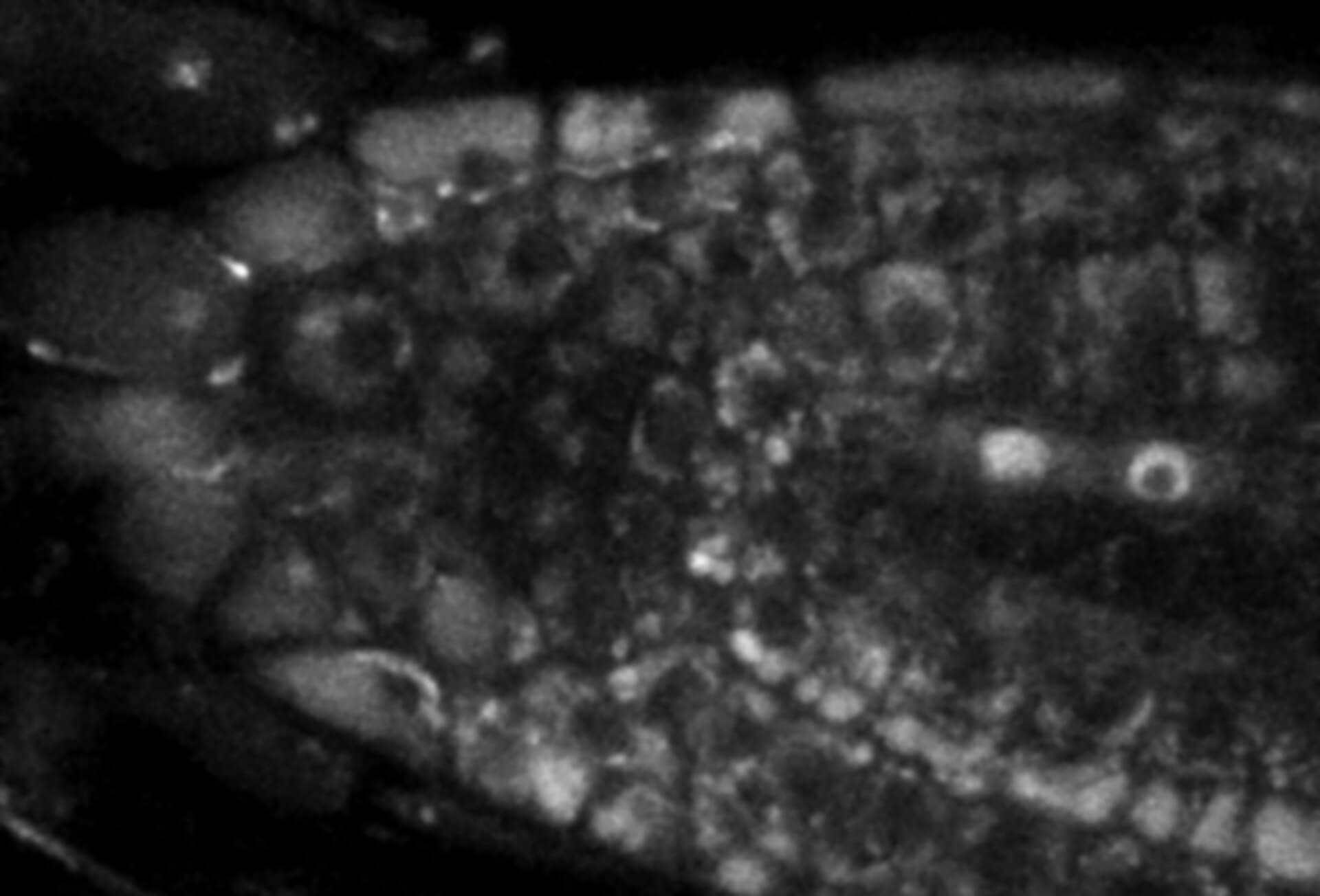
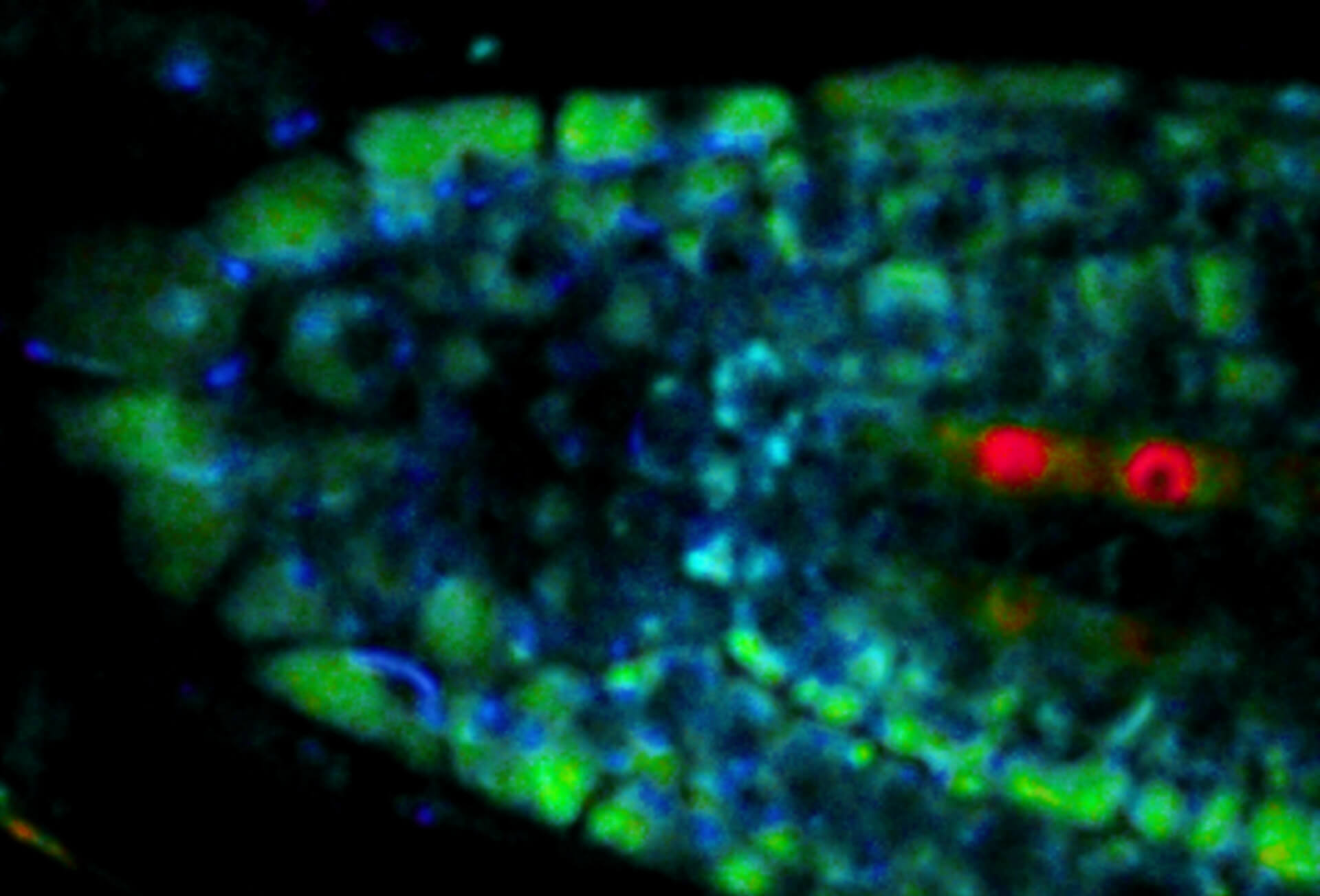
Movement of fluorophore-labeled 30S ribosomes (colored) in two E. coli bacteria (grayscale) tracked with MINFLUX.
Description
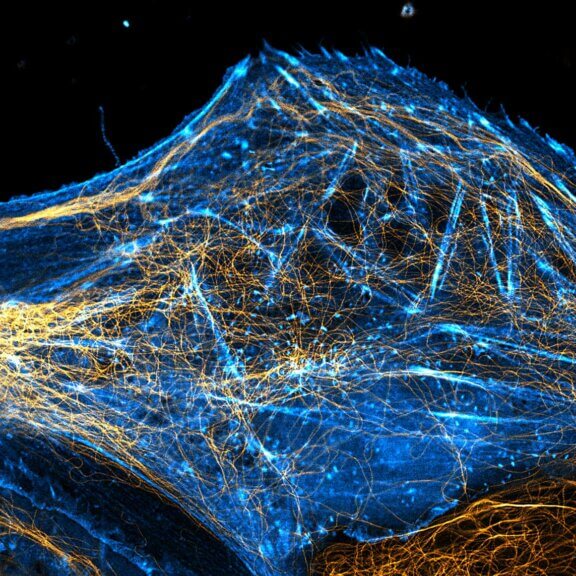
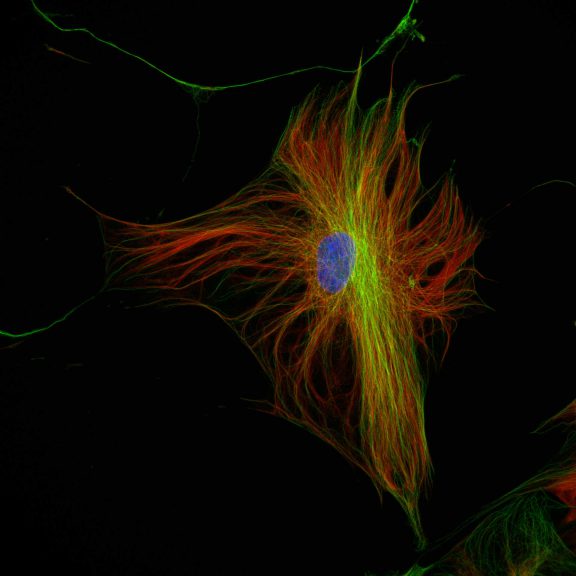
abberior STAR RED was coupled to polyclonal secondary nanobodies (alpaca VHH single domain antibodies) from Jackson ImmunoResearch and used to label tubulin in fixed mammalian cells via indirect immunofluorescence. The STED image was acquired with the STEDYCON system.
Description
Proteins of the nuclear pore complex and the golgi apparatus were stained by indirect immunofluorescence using abberior STAR RED (nuclear pore complex, magenta) and abberior STAR 580 (golgi, green) coupled to polyclonal secondary nanobodies (alpaca VHH single domain antibodies) from Jackson ImmunoResearch. Images of the fixed mammalian cells were acquired with the STEDYCON.
Description
Vimentin and tubulin were stained by indirect immunofluorescence using abberior STAR RED (vimentin, orange) and abberior STAR 580 (tubulin, cyan) coupled to polyclonal secondary nanobodies (alpaca VHH single domain antibodies) from Jackson ImmunoResearch. Images of the fixed mammalian cells were acquired with the STEDYCON.
Description
Indirect immunofluorescence staining with abberior STAR dyes coupled to polyclonal anti-mouse secondary nanobodies (alpaca VHH single domain antibodies) from Jackson ImmunoResearch shows excellent results in 3-color STED imaging. Staining was performed according to the premix & stain protocol. Vimentin (abberior STAR RED, green), tubulin (abberior STAR 580, magenta) and dsDNA (abberior STAR 460L, blue) were labeled in fixed mammalian cells and visualized with the STEDYCON.
Description
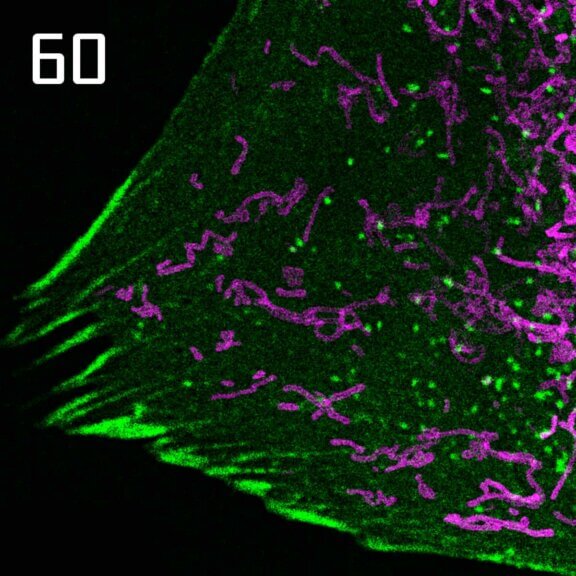
OMP25-SNAP protein in mitochondria labeled with abberior LIVE 610 SNAP (magenta) and Actin^K118TAG labeled with abberior LIVE 550 click (green). Movie with 60 frames.
Description
Description
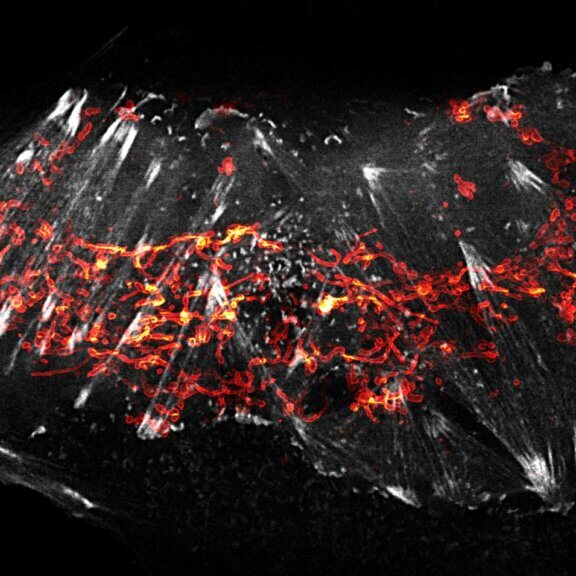
2-color STED image of a living cell expressing actin K118TAG incorporating TCO*A labeled with abberior LIVE 590 click (green). Plasma membrane is highlighted with abberior STAR RED membrane (magenta).
Description
2-color STED image of a living cell expressing actinK118TAG incorporating TCO*A labeled with abberior LIVE 550 click (cyan). Tubulin filaments were stained with our direct probe abberior LIVE 610 tubulin (yellow).
Description
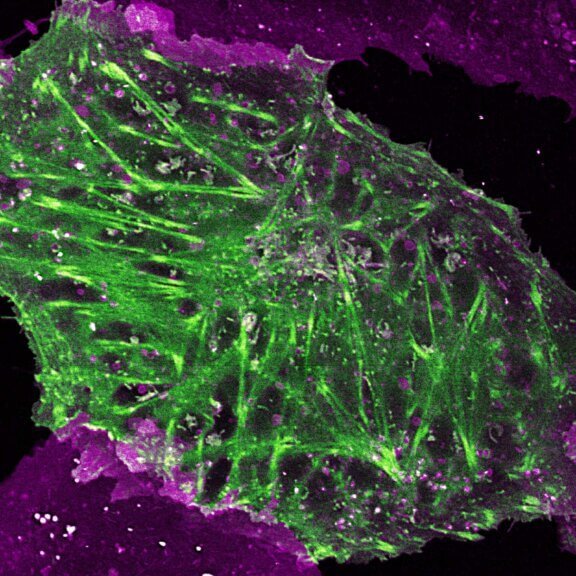
Perfect 3-color combination for STED @ 775 nm. abberior STAR RED NUP98 (red) and STAR ORANGE vimentin (cyan) complemented by the new STAR 460L tubulin (yellow). Cultured mammalian cells were fixed and labeled by indirect immunofluorescence.
Description
We are happy to introduce the first commercial long Stokes-shift dye for live-cell imaging applications. The NEW abberior LIVE 460L makes 3-color live-cell STED at 775 nm possible!
Live-cell STED image of a cell expressing a SNAP-tag® in an outer mitochondrial membrane protein visualized by our abberior LIVE 460L SNAP (orange). Tubulin filaments are highlighted with LIVE 610 (cyan) and the DNA with LIVE 560 DNA (red).
Description
STED time-lapse of living mammalian cell stained with abberior STAR ORANGE membrane.
Description
Two color live-cell STED and confocal image of a mammalian cultured cell stained with abberior STAR RED membrane (orange) and abberior LIVE 590 actin (cyan).
Description
Comparison of STED and confocal of a living mammalian cell stained with abberior STAR ORANGE membrane.
Description
Comparison of STED and confocal of a living mammalian cell stained with abberior STAR 488 membrane. Images were acquired with our FACILITY and STED @ 595 nm.
Description
Confocal time-lapse of living mammalian cell stained with abberior STAR 488 membrane.
Description
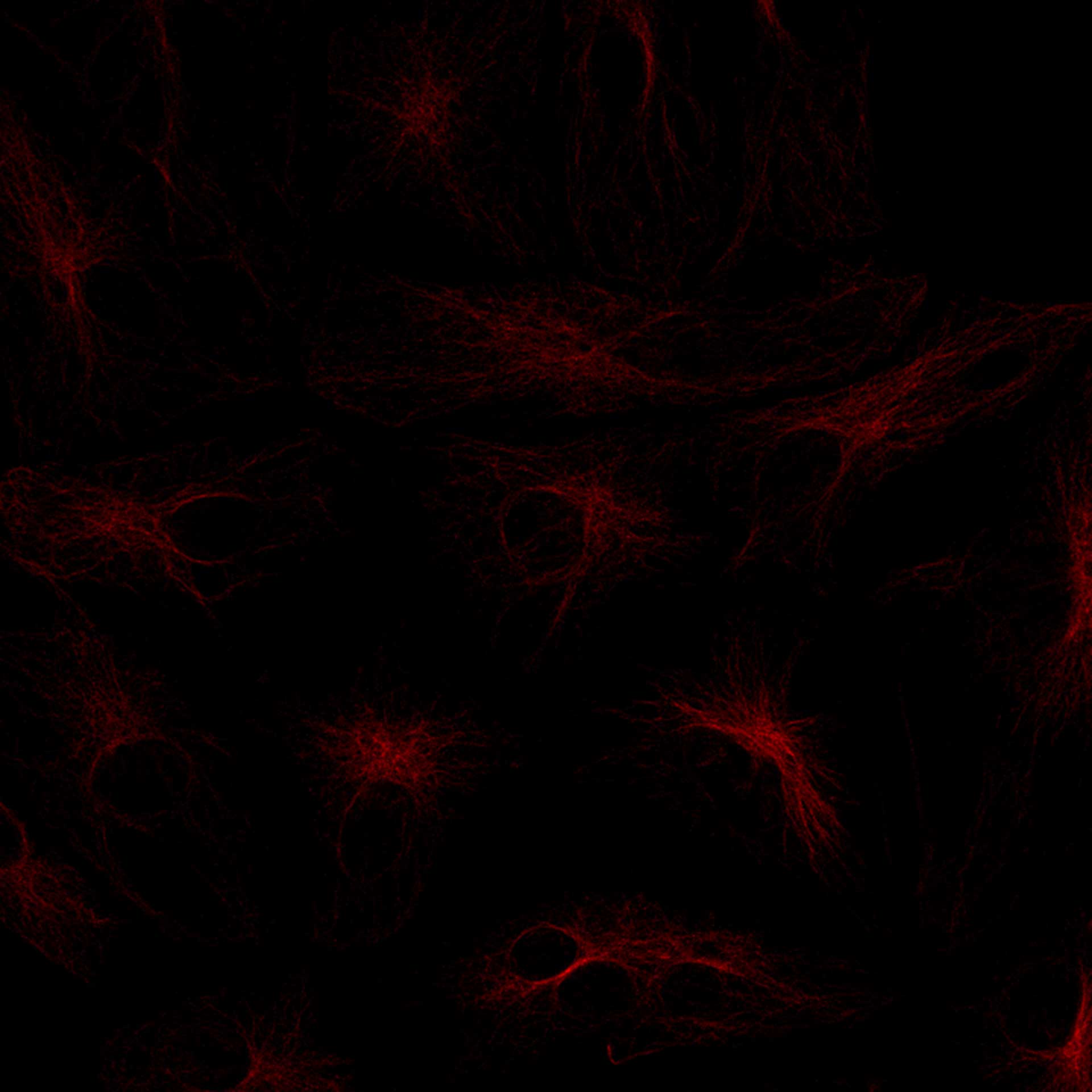
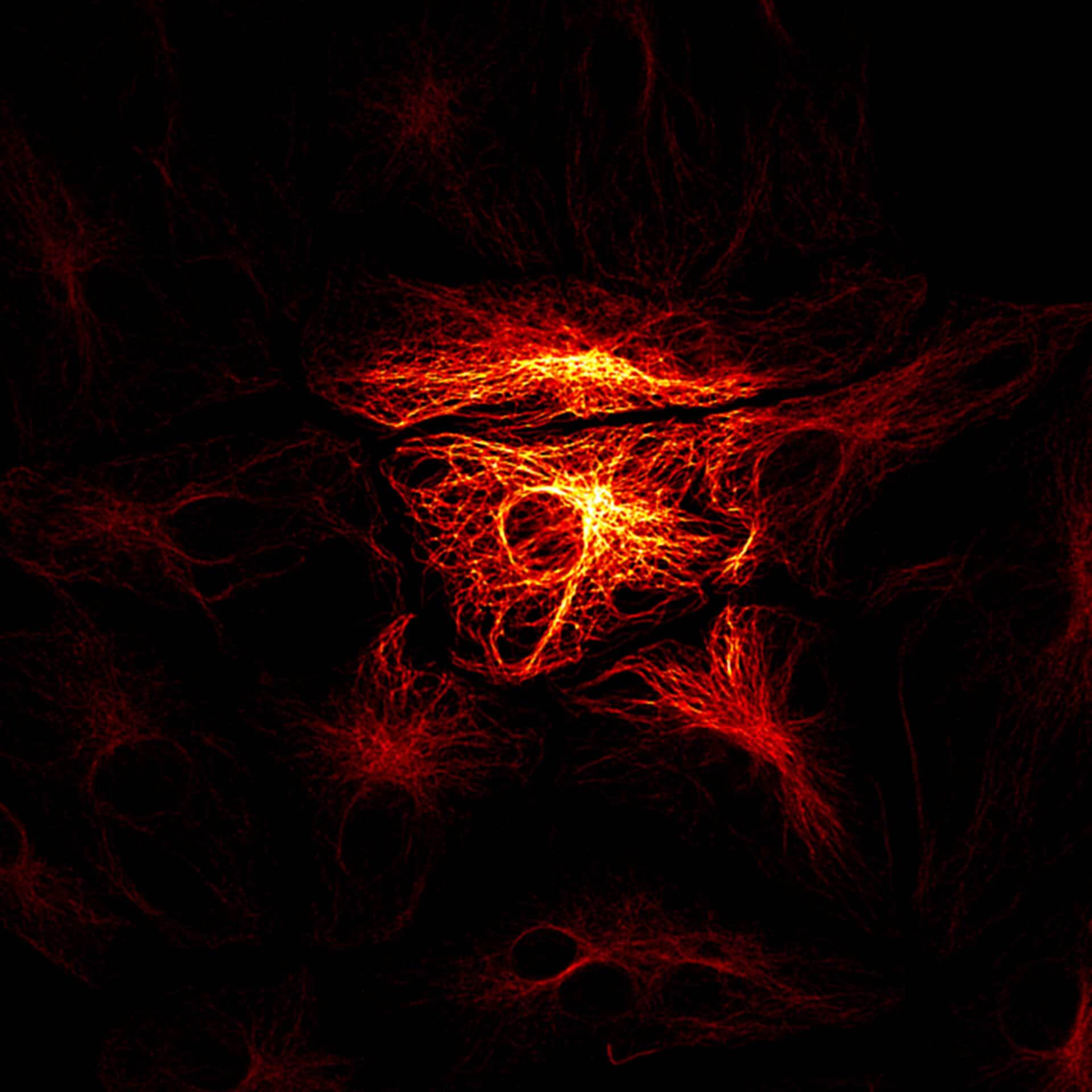
abberior CAGE 552 is a caged dye which is initially nonfluorescent and colorless substance which, upon photolysis with UV light, readily transforms irreversibly into a red and highly fluorescent dye.
The cytoskeleton is visible after UV induced uncaging of abberior CAGE 552.
Description
3D MINFLUX image of the mitochondrial import receptor Tom20 labeled with abberior FLUX 647 in fixed mammalian cells using indirect immunolabeling. Please note that Tom20 is only localized at the mitochondrial surface. The axial coordinate is color-coded.
Description
Two-color MINFLUX revealing an inner and outer mitochondrial membrane marker. Cultured mammalian cell labeled with indirect immunofluorescence using secondary antibodies coupled to abberior FLUX 640 (orange) and FLUX 680 (cyan). MINFLUX enables the visualization and separation of both structures.
Description
MINFLUX imaging of βII spectrin in a primary hippocampal neuron labeled with abberior FLUX 680 by indirect immunofluorescence. Please note the periodic arrangement of spectrin along the axon.
Description
MINFLUX image of axonal βII spectrin labeled with abberior FLUX 660 in primary hippocampal neurons. Note the periodic arrangement of spectrin along the axon, and the absence of any details in the confocal counterpart image.
Description
2D MINFLUX nanoscopy of the nuclear pore complex subunits. Fixed mammalian cells expressing SNAP-tag® NUP96 were labeled with abberior FLUX 647 SNAP. In contrast to confocal microscopy, 2D MINFLUX allows to visualize the shape and arrangement of individual subunits of the nuclear pore complex.
Description
2D MINFLUX imaging of the peroxisomal membrane protein PMP70 labeled with abberior FLUX 640 in fixed mammalian cells using indirect immunofluorescence.
Description
Drosophila ovariole stained with abberior LIVE 560 DNA showing nuclei in different cell types of the egg chamber. Ovaries were dissected from adult female fruit flies and were fixed prior to staining.
Image was acquired with the STEDYCON tiling feature and assembled with the SVI Huygens Stitcher.
Description
Drosophila spermatid tails were stained with abberior LIVE 610 tubulin. Testis were dissected from adult male fruit flies. Live cell imaging experiment was performed on a INFINITY microscope.
Description
Composite of three color live-cell image of an adherent mammalian cell. This living cell was directly labelled with abberior LIVE 510 actin (blue/green), LIVE 560 DNA (gray) and LIVE 610 tubulin. This image was acquired with a STEDYCON microscope.
Description
Three color live-cell STED at 775 nm: living cell labelled with abberior LIVE 460L (ER, green), LIVE 560 tubulin (magenta) and LIVE 610 actin (red).
Description
Confocal and STED time lapse of a living cell expressing SNAP-tag® fusion protein fusion protein in the endoplasmic reticulum lumen with SNAP and C-terminal tetrapeptide KDEL. SNAP-KDEL was labelled with abberior LIVE 610 SNAP ligand (benzylguanine).
The SNAP-KDEL plasmid was a gift from Francesca Bottanelli, FU Berlin.
Description
Live cell time lapse of abberior LIVE 590 DNA probe in cultured mammalian cells.
The use of very low concentrations of our abberior LIVE dyes reduces toxicity and allows long-term imaging of dynamic processes.
Image was acquired with the FACILITY microscope.
Description
Two color live-cell confocal and STED image of a mammalian cell expressing a SNAP-tag® OMP25 fusion protein decoration the outer membrane of mitochondria. OMP25 is visualized by our new abberior LIVE 610 SNAP ligand (orange). Tubulin filaments are highlighted with abberior LIVE 550 tubulin (cyan).
The SNAP-OMP25 plasmid was a gift from Francesca Bottanelli, FU Berlin.
Description
Two color live-cell confocal and STED image of a mammalian cell expressing a SNAP-tag® OMP25 fusion protein decoration the outer membrane of mitochondria. OMP25 is visualized by our new abberior LIVE 610 SNAP ligand (orange). Tubulin filaments are highlighted with abberior LIVE 550 tubulin (cyan).
The SNAP-OMP25 plasmid was a gift from Francesca Bottanelli, FU Berlin.
Description
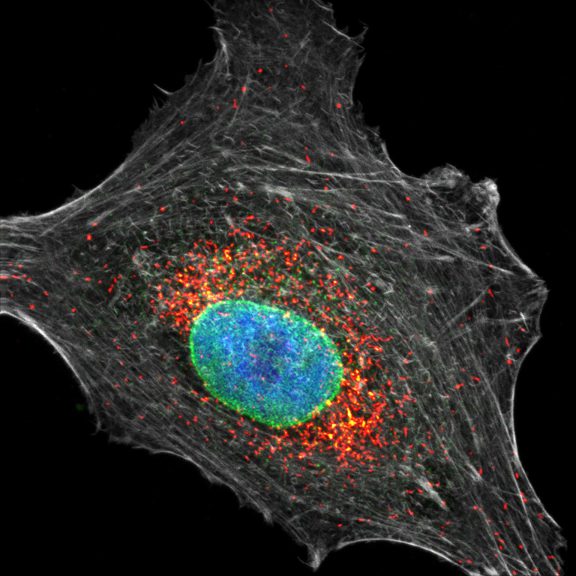
Cultured mammalian cell immunostained for an inner and outer mitochondrial membrane marker. Outer membrane is highlighted with abberior STAR RED (red) and the inner membrane with abberior STAR ORANGE (gray).
STED and confocal images were acquired with abberior's FACILITY microscope.
Description
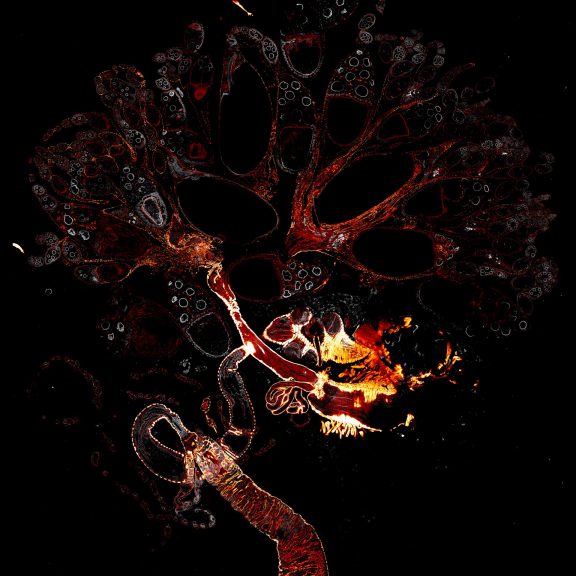
Drosophila male accessory gland stained for F-actin using abberior STAR 580 phalloidin.
Sample was prepared in cooperation with Dr. H. R. Shcherbata at MPl for Biophysical Chemistry, Göttingen, Germany.
Description
Three color STED and confocal image of a mammalian cultured cell immunostained for a nuclear pore protein in green and two golgi apparatus markers in red and blue.
abberior STAR RED highlights the golgi apparatus protein giantin (red) and abberior STAR ORANGE visualizes the cis-golgi protein GM130 (blue). NUP98 proteins were stained with abberior STAR GREEN (green).
Description
Three color confocal image of human fibroblast immunostained with abberior STAR 580 for vimentin (green) and with abberior STAR RED for tubulin (red).
DAPI was added to highlight the nucleus (blue).
Description
Four color confocal image of mammalian cultured cells. Peroxisomes were immunostained with abberior STAR RED (magenta) and nuclear pore proteins with abberior STAR 580 (green). F-actin fibers were highlighted with abberior STAR 488 phalloidin (gray).
The nucleus was visualized with DAPI (blue).
Description
Drosophila female reproductive system stained for F-actin (red) with abberior STAR RED phalloidin. abberior STAR ORANGE is highlighting a nuclear pore protein (gray).
Image was acquired with the STEDYCON tiling feature and assembled with the SVI Huygens Stitcher.
Description
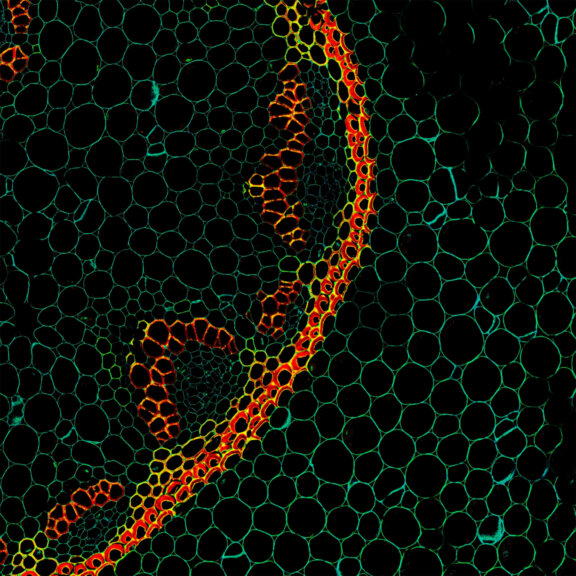
Autofluorescence in a plant stem cross section of Convallaria recorded with FACILITY. A large area, 10 by 10 tiles, each 125 µm by 125 µm, was imaged and stitched with image tiling in Fiji for ImageJ. This is combined with TIMEBOW lifetime imaging to show the differences in fluorescence lifetime due to the type of autofluorescent molecule and their nano-environment.
Modules:
Description
Live-cell sample of an Arabidopsis root tip suspended in water recorded with FACILITY. A subset of cells expresses a YFP construct. Lifetime imaging with TIMEBOW shows shifts in YFP fluorescence lifetime caused by the proteins nano-environment.
We thank Dr. Fábián Attila and Soós Vilmos (ATK, Brunszvik, HU) for providing this sample. Contact: soos.vilmos@atk.hu.
Modules:
Description
Autofluorescence in a plant stem cross section of Convallaria recorded with FACILITY. Image quality is improved by removing the background through MATRIX detection. This is combined with TIMEBOW lifetime imaging to show the differences in fluorescence lifetime due to the type of autofluorescent molecule and their nano-environment.
Modules:
Description
TIMEBOW STED vs MATRIX + TIMEBOW STED
Actin in microvilli of CaCo2 cells recorded with FACILITY. Fixed cells were stained with abberior STAR RED phalloidin. Imaging of the intense-background sample is optimized by MATRIX detection by directly subtracting unfocussed data. To boost resolution TIMEBOW is used, keeping STED laser power low. Lifetime information enables the selective exclusion of STED-affected emission at the rim of each measurement.
We thank Prof. Dorothee Günzel and Jörg Piontek (Charité, Berlin, DE) for providing this sample.
Modules:
Description
Confocal vs MATRIX + TIMEBOW STED
Actin in microvilli of CaCo2 cells recorded with FACILITY. Fixed cells were stained with abberior STAR RED phalloidin. Imaging of the intense-background sample is optimized by MATRIX detection by directly subtracting unfocussed data. To boost resolution TIMEBOW is used, keeping STED laser power low. Lifetime information enables the selective exclusion of STED-affected emission at the rim of each measurement.
We thank Prof. Dorothee Günzel and Jörg Piontek (Charité, Berlin, DE) for providing this sample.
Modules:
Description
MINFLUX 3D imaging of βII spectrin in a primary hippocampal neuron labeled with Alexa Fluor 647 by indirect immunofluorescence. The axial coordinate is color-coded. Please note the periodic arrangement of spectrin along the axon.
Description
MINFLUX 3D nanoscopy of nuclear pore complex subunits. 3D MINFLUX facilitates 3D imaging at molecular scales. In the axial direction localization, we reach precisions of approx. 2.5 nm in raw localization data. Labeling of NUP96-SNAP was performed using SNAP-Alexa Fluor 647.
Description
3D MINFLUX imaging of the peroxisomal membrane protein PMP70 labelled with abberior FLUX 647 in fixed mammalian cells using indirect immunofluorescence. 3D MINFLUX allows visualizing the shape of peroxisomes in all directions.
Description
MINFLUX 3D imaging of Clathrin coated vesicles and Clathrin coated pits. Labeling was performed using clathrin light chain-SNAP together with SNAP-Alexa Fluor 647.